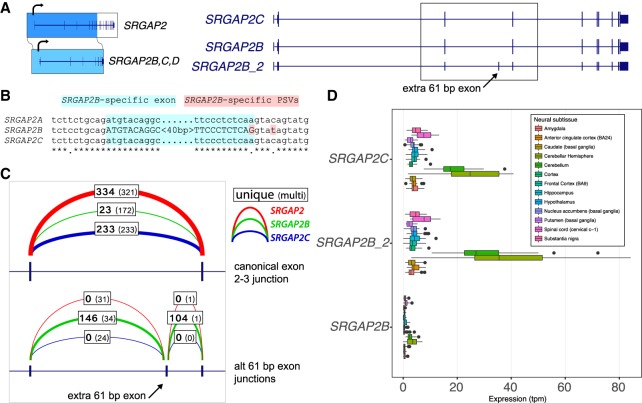
Figure 6.
Inclusion of a 61-bp exon and premature stop codon in SRGAP2B. (A) SRGAP2B and SRGAP2C are duplicate copies of SRGAP2, created by an initial 3′-truncated duplication ∼3 Myr ago. From the long-read capture data, we identify a major isoform (here SRGAP2B_2) containing an additional 61-bp exon (arrow). (B) Alignment of the 61-bp exon (highlighted in light blue) and flanking sequence identifies a key nucleotide change in the −1 position of the splice donor (highlighted in red). This A-to-G transition is predicted to substantially increase the strength of the splice donor signal. (C) Intron-spanning reads from GTEx RNA-seq data from brain (cortex) were counted for each paralog if they were consistent with the canonical exon 2–3 junction (above), or consistent with the extra 61-bp exon (below), with counts for SRGAP2 (red), SRGAP2B (green), and SRGAP2C (blue) shown. Boldface denotes uniquely mapping reads. The additional exon is frequently included in SRGAP2B transcripts, while rarely included if at all in SRGAP2A and SRGAP2C. (D) Expression estimates for isoforms shown in A generated using Kallisto and GTEx RNA-seq data corroborate that the isoform of SRGAP2B that includes the 61-bp exon is the predominant one in neural tissues. These subsequent mutational changes likely nonfunctionalized SRGAP2B and are consistent with the fixation of the granddaughter duplicate SRGAP2C in the human population.