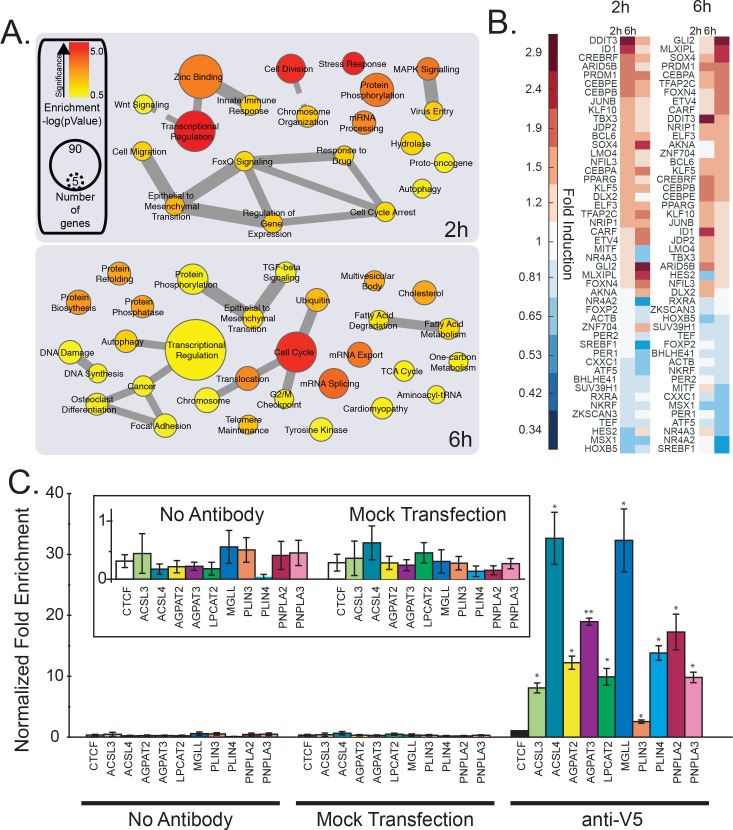
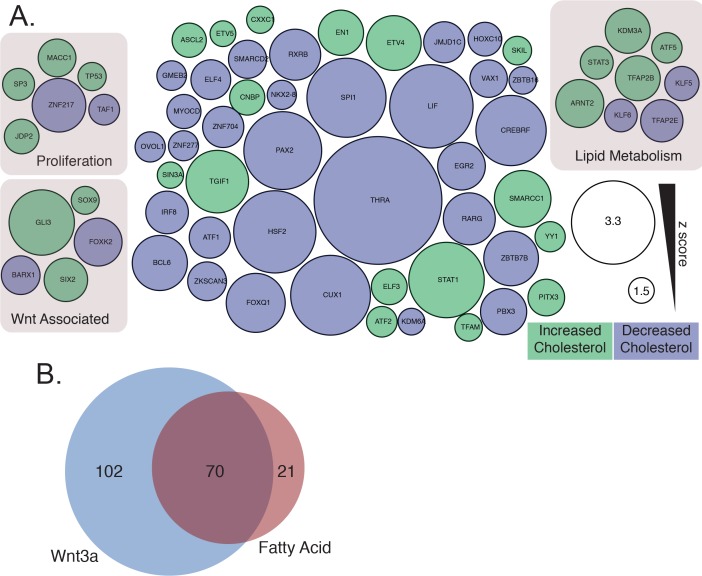
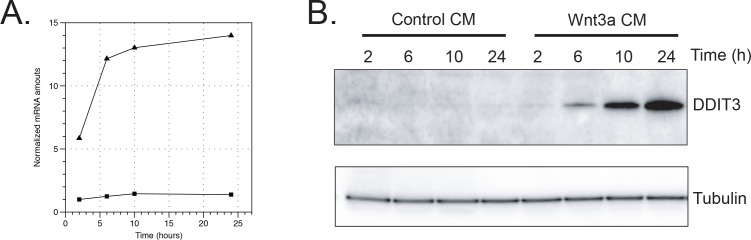
Figure 2. mRNA profiling and analysis of gene expression of cells treated with Wnt3a.
(A-B) HeLa-MZ cells were treated with control- or Wnt3a-conditioned media for 2 hr or 6 hr before RNA isolation and RNAseq analysis. Panel (A) shows the pathway enrichment of perturbed mRNAs. Node size indicates number of genes in each ontology and colour the statistical strength of the enrichment. Edge thickness indicates the strength of overlap of related ontologies. From (A), the fold change of transcription factors amounts in response to Wnt3a is shown in panel B. C. The ability of TFAP2 family member TFAP2A to bind to regulatory regions of lipid droplet at lipid metabolic enzyme genes was tested by ChIP-qPCR (see Materials and methods). Data are presented as the mean DNA amounts normalized to the negative control (CTCF) of three independent experiments ± SEM. (*) indicates a p-value<0.05; (**) indicates a p-value<0.005. Inset; re-scaled view of signal of the control conditions.