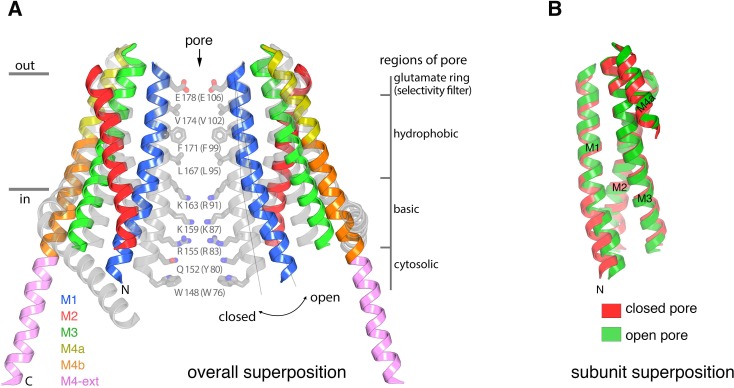
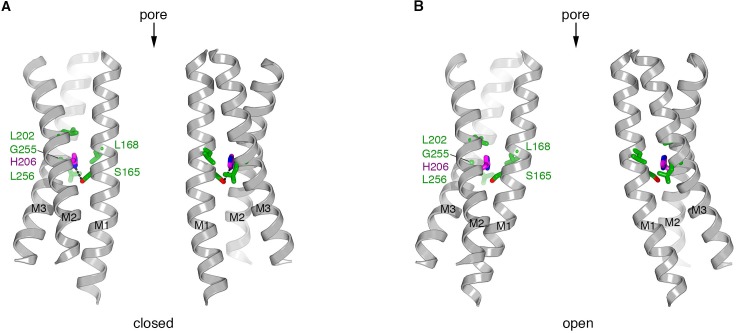
Figure 5. Conformational changes between the quiescent and open conformations.
(a) Superimposed structures of the quiescent (PDB ID: 4HKR) and open (H206A Oraicryst) conformations are drawn in ribbon representation. Two opposing subunits are shown, surrounding the pore, with the open conformation colored as indicated and the closed conformation in gray. Thin lines and a curved arrow highlight the outward rotation of subunits (with its fulcrum near Glu178) and the slight additional bend in M1. Conformational changes of M4/M4-ext are also apparent. Amino acids forming the walls of the closed pore (from the quiescent conformation) are shown as sticks, with corresponding regions of the pore indicated. Amino acids in parentheses denote human Orai1 counterparts. Horizontal bars indicate approximate boundaries of the plasma membrane. (b) Comparison of M1-M4a from individual subunits between the quiescent and open conformations. The region of an Orai subunit spanning M1 through M4a was superimposed between the quiescent (red ribbons, PDB ID 4HKR) and open (green ribbons, H206A Oraicryst) conformations. The slight additional bend in M1 of the open conformation is apparent at its N-terminal end. Otherwise the M1-to-M4b region of the two subunits superimpose within the error of the coordinates of the open conformation (the root-mean-squared deviation for the Cα positions of residues 163 to 288 superimposed in this manner is 1.1 Å).