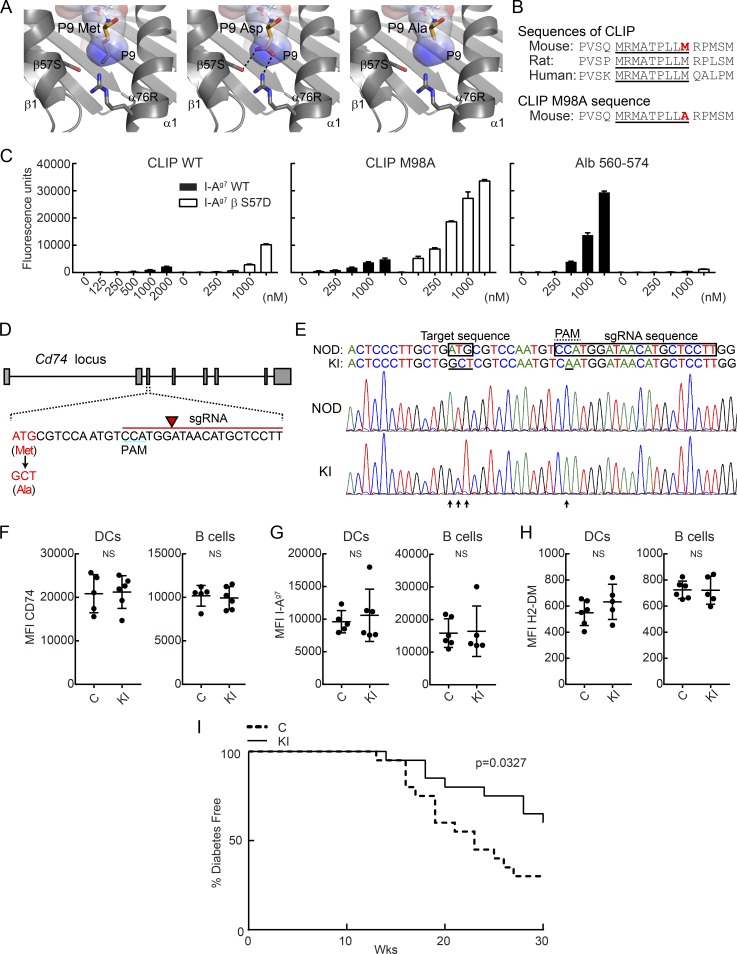
Figure 1.
Knock-in mutation in CLIP segment of invariant chain reduces incidence of T1D in NOD mice. (A) P9 pocket of I-Ag7–peptide complex. P9 Met residue of I-Ag7–GAD65 complex (left, PDB accession no. 1ES0) was mutated to Asp (middle) or Ala (right). P9 pocket is shown as a surface colored by electrostatic potential (blue = positive, red = negative). (B) Amino acid sequence of CLIP and CLIP M98A, highlighting the core sequence required for MHC II binding (underlined) and the P9 anchor residue (red). (C) Binding of peptides to soluble I-Ag7 WT or βS57D mutant protein. Indicated biotinylated peptides (125–2,000 nM) were incubated with soluble I-Ag7 proteins, and bound peptide was quantified by ELISA for triplicate samples. Mean + SD are shown. (D) sgRNA design used to introduce CLIP M98A mutation in the Cd74 gene. The sgRNA sequence is indicated in red, with arrow highlighting the site of DNA cleavage. (E) Nucleotide sequence of WT and M98A knock-in allele. The introduced mutation (ATG→GCT) is indicated as well as the silent mutation in the PAM (C→A). Chromatograms are shown for WT NOD and M98A/M98A (KI) mice. (F–H) Protein expression level of CD74 (F), I-Ag7 (G), and H2-DM (H) by splenic CD45+ I-Ag7+ CD11c+ CD11b− DCs and CD19+ I-Ag7+ B cells. Results are shown as mean ± SD with five to six mice/group. MFI, mean fluorescence intensity. (I) Diabetes incidence in control mice (C; n = 20) and cohoused M98A/M98A mice (KI; n = 20). Control mice were bred from same founders as M98A/M98A mice. Data are representative of two (C and F–H) or three independent experiments (I). Statistical analysis was performed with Mann-Whitney (F–H) or Gehan-Breslow-Wilcoxon test (I).