Abstract
Six endophytic bacteria were isolated from Saccharum sp (sugarcane) grown in the parish of Westmoreland on the island of Jamaica located in the West Indies. Whole genome sequence and annotation of the six bacteria show that three were from the genus Pseudomonas and the other three were from the genera Pantoea, Pseudocitrobacter, and Enterobacter. A scan of each genome using the antibiotics and secondary metabolite analysis shell (antiSMASH4.0) webserver showed evidence that the bacteria were able to produce a variety of secondary metabolites. In addition, we were able to show that one of the organisms, Enterobacter sp RIT418 produces N-acyl-homoserine lactones (AHLs), which is indicative of cell-cell communication via quorum sensing (QS).
Keywords: sugarcane, endophytes, quorum sensing, secondary metabolism
Introduction
Sugarcane is a tall perennial monocot from the genus Saccharum. The plant is native to warm temperate and tropical climates. The plant is agriculturally important because of sugar production. Sugarcane is an important crop plant in many countries including the island of Jamaica, where it is estimated that 100,000 metric tons of raw sugar will be produced from 1.4 million metric tons of sugarcane in 2018 1. Even though the plant is integral to the economy of Jamaica and other countries due it its role in sugar and ethanol production, studies related to endophytic and epiphytic bacterial-sugarcane interaction from sugarcane grown in Jamaica is very sparse.
Our laboratory is interested in assessing the bacterial-sugarcane symbiotic relationship for two main reasons. Firstly, to isolate and identify beneficial bacteria involved in nitrogen fixation. Secondly, to isolate and identify phyto-pathogens that are detrimental to the growth and/or development of the plant. A previous study from our group isolated and identified the bacterium Enterobacter sp strain SST3. Whole genome sequencing and annotation of Enterobacter sp strain SST3 show that the bacterium employs an AHL synthase gene involved in quorum sensing signaling. The AHL synthase gene from Enterobacter sp strain SST3 shares 88% similarity to the CroI gene from Citrobacter rodentium strain CC168 which is also involved in quorum sensing. In addition, Enterobacter sp strain SST3 possesses the complete genetic and proteomic machinery required for the catabolism of sucrose as an energy source in addition an indoleacetamide hydrolase (iaaH) ortholog involved in the production of auxin-like compound/s which has an integral role in plant growth and development 2. The work presented here is a continuation of the screening for additional beneficial and/or pathogenic endophytic bacteria. Here we present the isolation, genome sequencing and annotation of six endophytic bacteria isolated from the internal stem tissue of sugarcane grown in Jamaica. In addition, we present features of the isolates related to antibiotic production and other secondary metabolites such as; the production of compounds indicative of the quorum sensing cell-cell communication system.
Methods
Isolation of endophytic bacteria
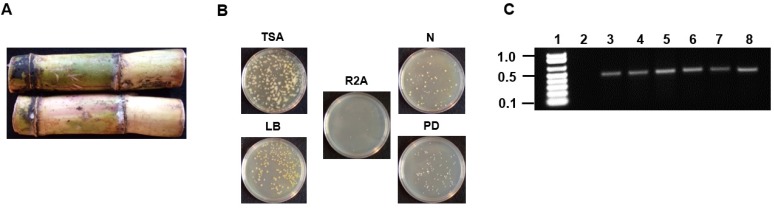
Sugarcane was obtained from a farm located in the parish of Westmoreland located on the island of Jamaica in the West Indies. The external surface of the sugarcane was sterilized using 1% (v/v) triton X-100 surfactant for 10 minutes followed by 20% (v/v) sodium hypochlorite/1% triton X-100 for 10 minutes followed by five 10 minutes washes using sterile distilled water. Following sterilization, the internal stem tissue was dissected under sterile conditions and 0.5 grams was used to inoculate 100 mL of 5 different media (tryptic soy, nutrient, R2A, Luria, and potato dextrose). The inoculated broth were allowed to incubate at 30oC for 48 hours with continuous shaking at 250 rpm. For isolation of pure colonies, serial dilutions (10-1 to 10-10) was performed and 100 μL of the samples ranging from 10-5 to 10-10 were plated on to the five different agar media (Fig. 1A and 1B).
Figure 1.
Isolation and PCR amplification of the V3/V4 16S rDNA region of endophytic bacterial isolates from Saccharum sp (A) Saccharum sp from Jamaica used for the isolation of endophytic bacteria. (B) Plating and isolation of bacteria on tryptic soy agar (TSA), Luria broth agar (LB), Reasoner's 2A agar (R2A), nutrient agar (N) and potato dextrose agar (PD). (C) 0.8% (w/v) agarose gel showing the resolution of the ~500bp amplicons of the V3/V4 16S rDNA region from the six endophytic bacteria. Lane 1-Marker (1Kb+), Lane 2-negative control (-DNA template), Lanes 3-8- V3/V4 amplicons from Pseudomonas sp RIT409, Pseudomonas sp RIT411, Pseudomonas sp RIT412, Pantoea sp RIT 413, Pseudocitrobacter sp RIT415, and Enterobacter sp RIT418 respectively.
N-acyl-homoserine lactone signal separation and detection
N-acyl-homoserine lactones (AHLs) were prepared and concentrated using ethyl acetate extraction of growth supernatants as previously describe by our laboratory 3, 4. T-streak, disc diffusion and thin layer chromatography (TLC) bioassays were done as described in previous original publications by our laboratory 3-5 and in corresponding review articles on biosensors for AHL detection 6, 7.
Genomic DNA isolation and PCR amplification of the 16S V3/V4 rDNA regions
Genomic DNA was extracted from 5 mLs of individual bacteria grown in broth using the MolBio DNA extraction kit according to the manufacturer's instructions. For initial identification of isolates, the variable 3 and 4 (V3/V4) regions of the 16S rDNA was amplified using 12 picomoles of forward and reverse primer, 1mM MgSO4, 0.5 mM of each of the four deoxynucleotide triphosphates, 0.2 ng genomic DNA and 1 unit of platinum Pfx DNA polymerase (Invitrogen) using the following PCR conditions: 1 cycle at 95oC for 2 minutes, followed by 25 cycles at 95oC for 30 seconds, 52oC for 30 seconds and 72oC for 1 minute. The forward and reverse primers used to amplify the V3/V4 region were 5'-CCTACGGGNGGCWGCAG-3' and 5'-GACTACHVGGGTATCTAATCC-3' (Fig. 1C). The ~500bp V3/ V4 amplicons were resolved by electrophoresis on s 0.8% (w/v) agarose gel followed by gel extraction using the QIAquick Gel Extraction Kit (Qiagen) followed by Sanger nucleotide sequencing in both directions using the primers that were used for amplification. The individual genera were identified using the Basic Local Alignment Search Tool (BLAST) 8.
Genome sequencing and assembly
For whole genome sequencing, the extracted DNA was processed using the Nextera XT (Illumina), quantified using a NanoDrop spectrophotometer and sequenced using the MiSeq Illumina platform at the Rochester Institute of Technology Genomics Facility. Adapter trimming was performed on the raw paired-end reads using SeqPurge version 0.1. The trimmed reads were subsequently assembled de novo with Unicycler version 0.3.0b.
Strain identification and genome quality assessment
For each assembled genome, 43 conserved microbial marker genes were identified, concatenated and used to determine strain identity based on phylogenetic placement within a reference genome tree consisting of 5,656 trusted reference genomes 9. Lineage-specific marker genes were subsequently inferred for each genome based on updated taxonomic assignment and was also used to estimate genome completeness and contamination. The taxonomic assignment of Pseudocitrobacter sp RIT415 to the family Enteobacteriaceae showed strikingly high 16S rDNA identity (>99.5%) to members from the genus Pseudocitrobacter. Since there were no reference published genomes from the genus as of June 28, 2018, a nucleotide BLAST search was done using its whole genome as the query against seven house-keeping genes (gyrB, rpoA, rpoB, trmE, recN, infB, atpD) of Pseudocitrobacter faecalis 25 CITT and Pseudocitrobacter anthropi C138T. The result of this comparison between the seven house-keeping genes of Pseudocitrobacter sp RIT415 and Pseudocitrobacter faecalis 25 CITT and Pseudocitrobacter anthropi C138T shows that >98% identity for each (data not shown).
Results and Discussion
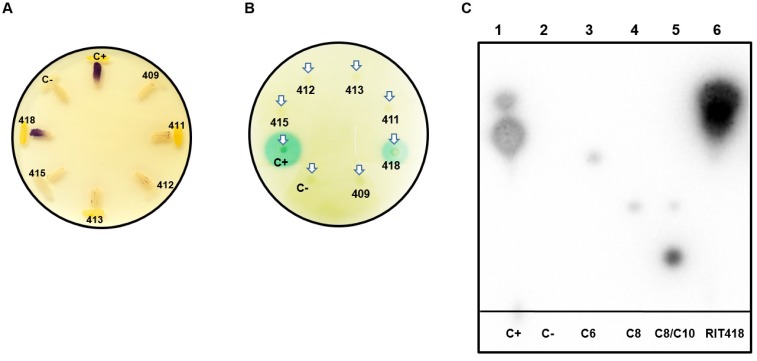
Approximately 500 MB of paired-end reads were generated from the whole genome sequencing for each of the six isolates. De novo genome assembly followed by CheckM inspection indicates that the assembled genomes are of good quality with high completeness (>98%) and negligible or possibly background contamination (<0.5%) (Table 1). CheckM, JSpecies and nucleotide BLAST analyses assigned the isolates to the genera Pseudomonas, Enterobacter, Pantoea or Pseudocitrobacter (Table 1). It is notable that this is the first reported genome representative for Pseudocitrobacter as of June 28, 2018. AntiSMASH results for the isolates in this study showed the production of various secondary metabolites, including antibiotics as listed in Table 2. Interestingly, an N-acyl homoserine lactone synthase (luxI) homolog was identified in the Pantoea sp RIT413 genome. However, the strain did not accumulate AHLs that could activate the receptors TraR or CviR proteins (Fig. 2). In contrast, a luxI homolog could not be identified in Enterobacter sp RIT418 using both BLAST and antiSMASH queries. However, the strain was able to produce TraR and CviR-detectable AHL signal/s (Table 2, Fig. 2). To be sure that this was not a result of human error during library preparation and culture deposition, the six isolated strains were re-streaked from our culture repository to assess purity. In addition, the V3/V4 region of the 16S rDNA were re-amplified, re-sequenced and analyzed. The reanalysis corroborated or our original findings. Nucleotide and protein BLAST searches was done using the croI/luxI and easI homologs from Enterobacter sp strain SST3 and Enterobacter asburiae stain L1 respectively as queries 2, 10. The BLAST searches did not identify any luxI or luxI-like homologs in Enterobacter sp RIT418. A complete genome of Enterobacter sp RIT418 can be obtained more readily given the recent advancement of Nanopore and PacBio long read technologies. Sequencing from Nanopore and PacBio will be instructive in corroborating the absence of luxI homlog(s) in the genome of Enterobacter sp RIT418 11. It would also be interesting to perform future experiments to identify and characterize the gene/s and proteins that are responsible for the production AHL signals in Enterobacter sp RIT418 using forward and or reverse genetic approaches. The lack of the TraR or CviR-detectable AHL signal(s) production in the luxI-ortholog-containing Pantoea sp RIT413 may be associated with the regulation of the luxI homolog. This will require recombinant cloning and heterologous expression of the luxI ortholog employing a broad host-range expression vector to detect AHL production 12. It is also possible that Pantoea sp RIT413 does accumulate AHLs but the compounds produced are structurally divergent from the typical TraR and CviR cognate AHL substrates which may not be able to activate luxR-type receptors.
Table 1.
Genome annotation information for the isolated strains. The isolated strains were identified to the genus level based on high nucleotide similarity (> 97%) against 16S rDNA and various house-keeping genes (gyrB, rpoA, rpoB, trmE, recN, infB, atpD) of species type strains.
| Organism | CheckM Marker Lineage | Accession no. | Genome Size (bp) | %GC Content | Genome Coverage (X) | No. of Contigs | N50 (contigs) | Contamination | Completeness |
|---|---|---|---|---|---|---|---|---|---|
| Pseudomonas sp RIT409 | Pseudomonas | QARE00000000 | 5918228 | 60.1 | 57 | 57 | 233947 | 0.14 | 99.68 |
| Pseudomonas sp RIT411 | Pseudomonas | QBIZ00000000 | 5310967 | 66.1 | 53 | 103 | 91721 | 0.22 | 99.67 |
| Pseudomonas sp RIT412 | Pseudomonas | QBJA00000000 | 5924148 | 60.1 | 80 | 48 | 355428 | 0.14 | 99.68 |
|
Pantoea sp RIT413 |
Pantoea | QBJB00000000 | 3836880 | 56.6 | 70 | 38 | 234727 | 0.33 | 99.28 |
| Pseudocitrobacter sp RIT415 | Enterobacteriacieae (Pseudocitrobactera) | QBJC00000000 | 4743521 | 53.5 | 80 | 52 | 164365 | 0.12 | 100 |
| Enterobacter sp RIT418 | Enterobacter | QBJD00000000 | 4557387 | 57.0 | 62 | 44 | 238708 | 0.11 | 98.83 |
Table 2.
antiSMASH analysis showing the presence/absence of genes clusters involved in the synthesis of antibiotics and secondary metabolites.
| Organism | Siderophore | NPRS | Thiopeptide | Terpene | Hserlactone | t2pks-arylpolyene | Bacteriocin | Arylpolyene | Acyl amino acids |
|---|---|---|---|---|---|---|---|---|---|
| Pseudomonas sp RIT409 | + | - | + | + | - | - | + | - | - |
| Pseudomonas sp RIT411 | + | + | - | + | - | - | - | + | - |
| Pseudomonas sp RIT412 | + | + | + | + | - | - | - | - | - |
|
Pantoea sp RIT413 |
+ | - | + | + | + | - | - | - | + |
| Pseudocitrobacter sp RIT415 | - | + | + | - | - | + | + | - | - |
| Enterobacter sp RIT418 | + | + | + | - | - | + | - | - | - |
Figure 2.
Detection of N-acyl-homoserine lactones (AHLs) quorum-sensing signals from sugarcane bacterial isolates. (A) Detection of AHLs with biosensor CV026 in a t-streak bioassay (B) Detection of AHLs with biosensor NTL4 (pZRL4) in a disc diffusion bioassay. (C) Separation of AHLs from isolate Enterobacter sp RIT418 using thin layer chromatography coupled to an NTL4 (pZRL4) agar overlay for AHL detection. Lane 1- positive control (C+) (ethyl acetate extract from Pantoea stewartii DC283), Lane 2- negative control (C-) (ethyl acetate extract from E. coli Dh5α), Lane 3-C6-HSL standard, Lane 4-C8-HSL standard, Lane 5-C8/C10-HSL standard and Lane 6-ethyl acetate extract from Enterobacter sp RIT418.
Nucleotide sequence accession numbers
The genome sequences of the strains described in this study have been deposited in the GenBank database with the accession numbers and annotation features describes in Table 1. The version described in this paper is the first version.
Acknowledgments
AOH, AP, MAS, NHW, KKS, and KRH acknowledge the Thomas H. Gosnell School of Life Sciences (GSoLS) and the College of Science (COS) at the Rochester Institute of Technology (RIT) for ongoing support. KKS and KRH are undergraduate research students in the laboratories of AOH and MAS respectively. We dedicate the work to the memory of our late colleague and friend, Dr. David A. Lawlor.
References
- 1.Grant C. USDA Gain Report (Jamaica Sugar Annual Report) USDA; 2017. [Google Scholar]
- 2.Gan HM, McGroty SE, Chew TH, Chan KG, Buckley LJ, Savka MA, Hudson AO. Whole-genome sequence of Enterobacter sp. strain SST3, an endophyte isolated from Jamaican sugarcane (Saccharum sp.) stalk tissue. J Bacteriol. 2012;194:5981–2. doi: 10.1128/JB.01469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gan HM, Buckley L, Szegedi E, Hudson AO, Savka MA. Identification of an rsh gene from a Novosphingobium sp. necessary for quorum-sensing signal accumulation. J Bacteriol. 2009;191:2551–60. doi: 10.1128/JB.01692-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott RA, Weil J, Le PT, Williams P, Fray RG, von Bodman SB, Savka MA. Long- and short-chain plant-produced bacterial N-acyl-homoserine lactones become components of phyllosphere, rhizosphere, and soil. Mol Plant Microbe Interact. 2006;19:227–39. doi: 10.1094/MPMI-19-0227. [DOI] [PubMed] [Google Scholar]
- 5.Lowe N, Gan HM, Chakravartty V, Scott R, Szegedi E, Burr TJ, Savka MA. Quorum-sensing signal production by Agrobacterium vitis strains and their tumor-inducing and tartrate-catabolic plasmids. FEMS Microbiol Lett. 2009;296:102–9. doi: 10.1111/j.1574-6968.2009.01627.x. [DOI] [PubMed] [Google Scholar]
- 6.Farrand SK, Qin Y, Oger P. Quorum-sensing system of Agrobacterium plasmids: analysis and utility. Methods Enzymol. 2002;358:452–84. doi: 10.1016/s0076-6879(02)58108-8. [DOI] [PubMed] [Google Scholar]
- 7.Steindler L, Venturi V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol Lett. 2007;266:1–9. doi: 10.1111/j.1574-6968.2006.00501.x. [DOI] [PubMed] [Google Scholar]
- 8.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Parks D, Imelfort M, Skennerton C, Hugenholtz P, Tyson G. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells and metagenomes. Genome Research. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau YY, Yin WF, Chan KG. Enterobacter asburiae strain L1: complete genome and whole genome optical mapping analysis of a quorum sensing bacterium. Sensors (Basel) 2014;14:13913–24. doi: 10.3390/s140813913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan HM, Lee YP, Austin CM. Nanopore Long-Read Guided Complete Genome Assembly of Hydrogenophaga intermedia, and Genomic Insights into 4-Aminobenzenesulfonate, p-Aminobenzoic Acid and Hydrogen Metabolism in the Genus Hydrogenophaga. Front. Microbiol. 2017;8:1880.. doi: 10.3389/fmicb.2017.01880. doi: 10.3389/fmicb.2017.01880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gan HM, Dailey LK, Halliday N, Williams P, Hudson AO, Savka MA. Genome sequencing-assisted identification and the first functional validation of N-acyl-homoserine-lactone synthases from the Sphingomonadaceae family. PeerJ. 2017;4:e2332. doi: 10.7717/peerj.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]