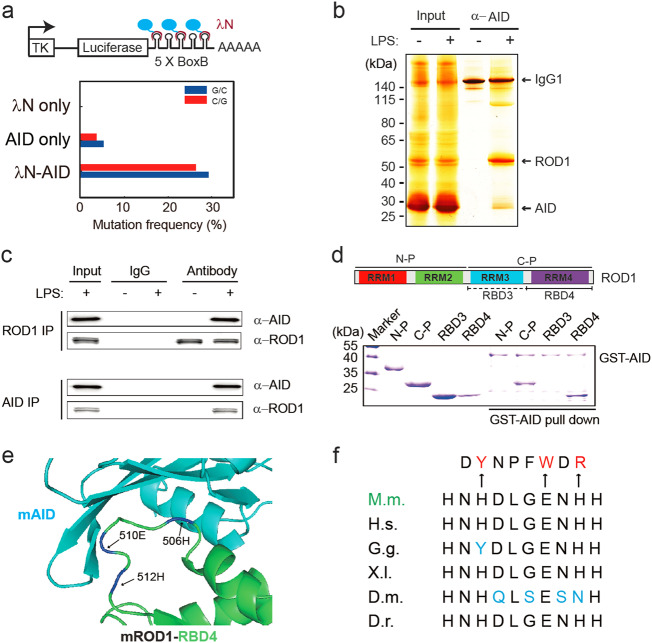
Fig. 1.
ROD1 physically interacts with AID via an ultraconserved loop region. a Diagram of the λN/BoxB tethering assay and the mutation frequency observed in HEK293 cells. The C/G mutations to all C/G bases in BoxB region were calculated from 20 sequenced clones. b Silver staining of AID immunoprecipitates from lysates of either LPS-activated or naive splenic B cells. c ROD1 and AID interact with each other in LPS-activated B cells. The reciprocal co-IP was probed with anti-AID and anti-ROD1 antibodies. d Direct interaction between AID and ROD1 truncated proteins by GST pull-down assay. RRM RNA recognition motif, N-P N-terminal protein, C-P C-terminal protein, RBD3 RNA-binding domain 3, RBD4 RNA-binding domain 4. e The 3D interacting surface of AID (cyan) and ROD1 (green) modeled by PRISM. The key interacting amino acids are labeled in blue and indicated by arrowheads. f The residue composition and conservation of the loop region in ROD1. Amino acids from 504 to 513 were aligned across the animal kingdom. The mutated amino acids at each position are listed and marked by arrowheads. D.r. zebrafish, D.m. fly, X.I. frog, G.g. chicken, H.s. human, M.m. mouse