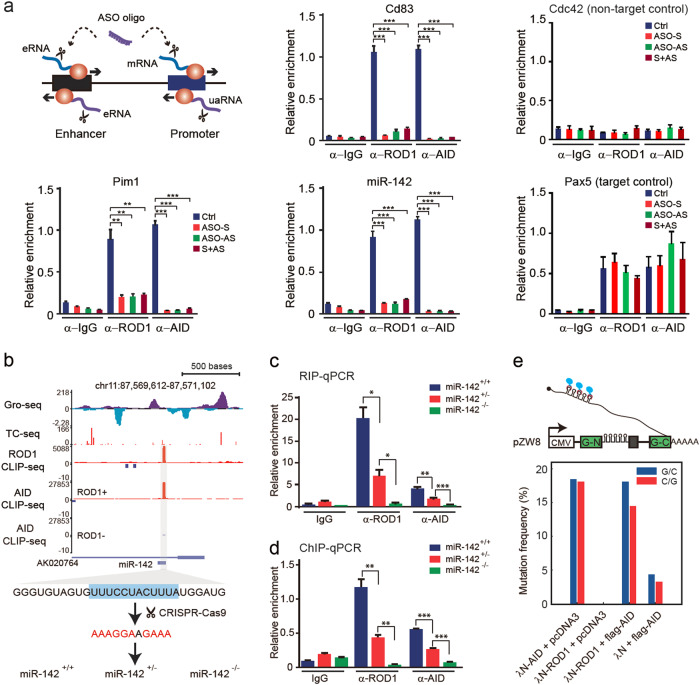
Fig. 6.
ROD1 cotranscriptionally recruits AID via bi-directional RNA. a ChIP-qPCR analysis of ROD1 and AID occupancy in AID target and non-target regions upon depletion of bi-directional RNA. Gene-specific ASOs to Cd83, Pim1, and miR-142 were individually transfected into LPS-activated B cells for 48 h. Cdc42 served as an AID non-target control, whereas Pax5 as AID target control, both of the samples were treated by CD83-ASO oligos. The relative enrichment was calculated by normalizing all of the data against input DNA. Data are presented as the mean ± SD (n = 3). The cartoon depicts bi-directional transcription at promoters and enhancers in mammalian cells. eRNA enhancer RNA, uaRNA promoter upstream antisense RNA. b Snapshot of the Gro-seq, TC-seq, and CLIP-seq profiles in the vicinity of the miR-142 locus. The ROD1-binding sequence highlighted in cyan was mutated into AG-rich sequences by CRISPR-Cas9 in CH12F3 cells. c, d RIP-qPCR or ChIP-qPCR analysis of ROD1 and AID occupancy at the miR-142 locus in different genotypes. The data are presented as the mean ± SD (n = 3). e Diagram of the λN/BoxB intronic tethering assay and the mutation frequency observed in HEK293 cells. The C/G mutation frequency was determined by Sanger sequencing from 20 sequenced clones. *P < 0.05, **P < 0.01 and ***P < 0.001 by two-tailed Student’s t test