Fig. 7.
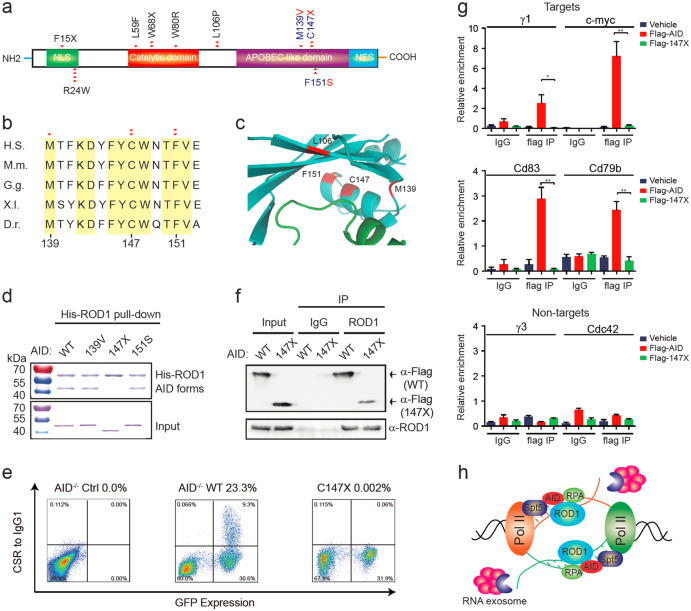
HIGM2 mutations in AID disrupt its interactions with ROD1. a Diagram of the domain structure of AID and HIGM2-related mutations. Naturally occurring mutations in patients (red triangle) from a previous report46 are shown at the exact residue positions. b The mutation residues are highly conserved. Amino acids from 139 to 153 were aligned. D.r. Zebrafish, X.l. frog, G.g. chicken, H.s. human, M.m. mouse. c The interacting surface between human AID and ROD1 modeled by PRISM. The mutations are marked in red. d His-ROD1 pulled-down GST-AID and some variant proteins, but not 147X variant. e Rescue of IgG1 CSR in AID−/− splenic B cells by transducing Flag-tagged AID or AID147X mutant. Percentage of IgG1 CSR is the ratio between IgG1+/GFP+ cells and total GFP+ cells. f Anti-ROD1 immunoprecipitates from retrovirally transduced AID−/− B cells with either Flag-tagged AID or AID147X mutant. g ChIP-qPCR analysis of AID and AID147X mutant occupancy at S-γ1 and non-Ig targets in reconstituted AID−/− B cells as shown in f. S-γ3 and Cdc42 served as AID non-target controls. *P < 0.05 and **P < 0.01 by two-tailed Student’s t test. The data are presented as the mean ± SD (n = 3). h A model of bi-directionally trapped ROD1 recruiting AID. In naive B cells, ROD1 resides on bi-directionally transcribed RNA and waits for the expression of AID. Once AID is dramatically induced upon antigen stimulation, it will be guided to the defined sites to initiate CSR, SHM and translocations