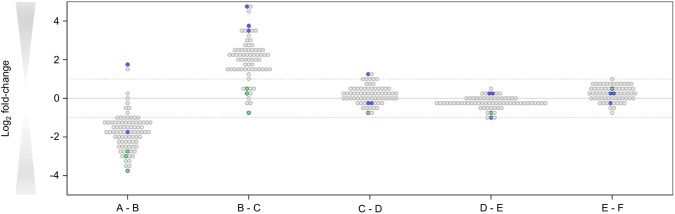
Figure 4.
Differences in absolute abundances of OTUs detected at consecutive locations within the DWDS. OTU-level differences between DWDS locations (A–F) were calculated by dividing the number of 16S rRNA gene copies per OTU derived from two consecutive locations. For this, the medians of 16S rRNA gene copies per OTU that were measured over a 5-month period were used and their differences between two consecutive locations plotted using a binary logarithmic scale. OTUs with a relative abundance of <5% per sampled location are shown as grey dots, whereas OTUs with >5% relative abundance at location A (Peribacterales, Parcubacteria, and Gallionellaceae) are shown as green dots and OTUs with >5% relative abundance at location F (Comamonadaceae, Deferrisoma, and Nitrospira) are shown as blue dots. Differences with more than a 2-fold increase/decrease (dotted lines) were considered as significant differences that cannot be explained by technical variations introduced during sample processing.