Figure 1.
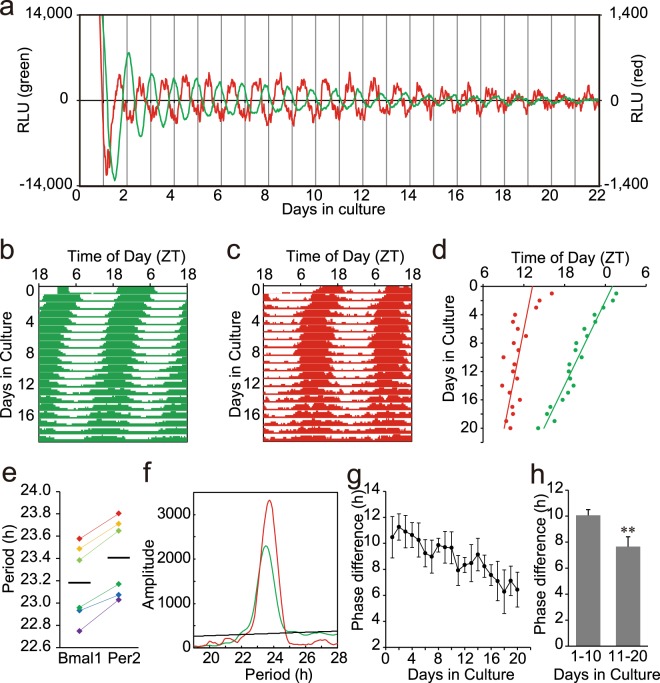
Circadian rhythms in the neonatal SCN slices of Bmal1-Eluc:Per2-SLR2 mice (a) Sequential records of circadian Bmal1 (green) and Per2 (red) rhythm in a neonatal SCN slice. Original records were smoothed by a 5 point (80 minutes) moving average method and detrended by a 24 hour moving subtraction method. The abscissa indicates the day in culture and the ordinate the relative intensity of bioluminescence (right, Bmal1; left, Per2). Day 0 means the day of slice preparation. (b,c) A double plotted circadian Bmal1 (b) and Per2 (c) rhythm in a raster format. Bioluminescence data of each day were indicated as a percentile to the daily mean. (d) Successive plots of circadian peak phases (Bmal1, green; Per2, red) and regressions lines fitted to them in the same SCN slice as used in (a). (e) Circadian periods of individual Bmal1 and Per2 rhythms (n = 6) calculated from a regression line. A horizontal bar indicates the mean circadian period. Circadian periods of the rhythms from the same SCN are indicated with the same colors. (f) Chi-square periodogram demonstrates significantly different circadian periods of Bmal1 (green) and Per2 (red) rhythm. A black oblique line in the graph indicates the significance level at p < 0.05. (g) Phase difference (ψ) between the circadian Bmal1 and Per2 peaks in each cycle. The values are indicated as the mean and SD. (h) The mean and SD of ψ in the first and the last 10 cycles is indicated. **p < 0.01 (paired t-test).