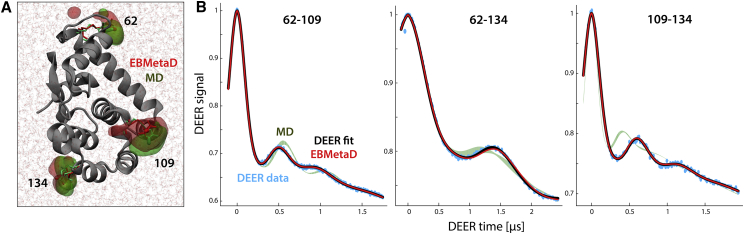
Figure 10.
Evaluation of generalized EBMetaD for spin-labeled T4L. (A) A closeup of T4L in the MD simulation system is given, highlighting the spin labels at positions 62, 109, and 134. Red and green surfaces encompass the regions occupied by the nitroxide groups during the EBMetaD and MD simulations, respectively. The root mean-square deviation of the protein backbone (gray cartoons) relative to the initial x-ray structure (46) is within 2 Å in both cases. (B) For each of the three spin-labeled pairs, the experimental DEER signals (cyan) and the corresponding fits (black) are compared with theoretical signals calculated (Eqs. 2, 3, 4, 5, and 6) from the simulated EBMetaD distributions (red) in Fig. 9B, i.e., in consideration of the confidence bands. The calculated signals from the unbiased MD data are also shown for comparison (green bands). The Δ and λ parameters for each of these calculated time-traces are provided in Table S7.