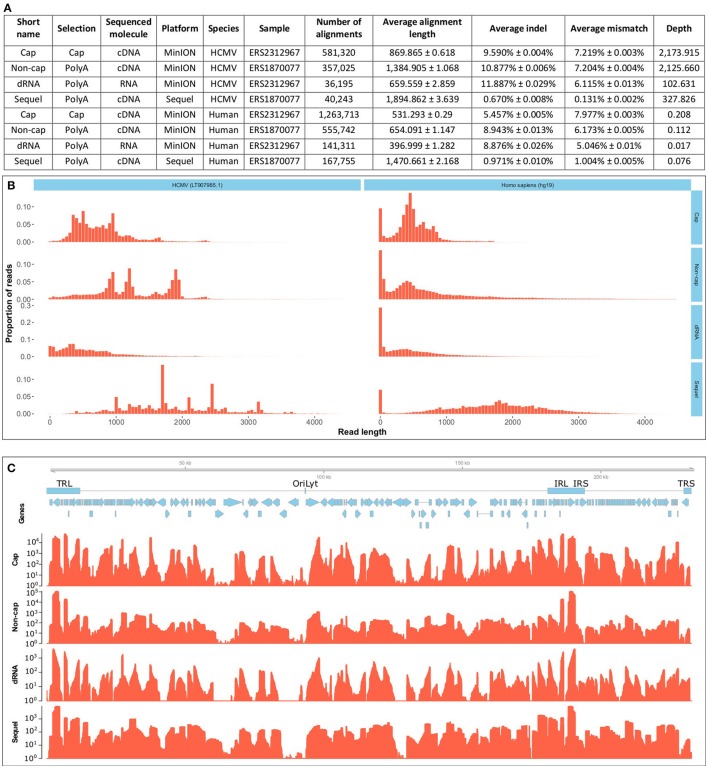
Figure 1.
Data quality and metrics. (A) Summarizes the quality metrics of the sequencing reads for each sequencing run broken down per species (host and virus). All errors are given as standard errors. (B) Depicts the read-length distribution. The files have been uploaded to the ENA under the study accession PRJEB25680. Aligned read length is expressed in base pairs and the distribution is depicted for 50-basepair-long bins. The x axis is only presented until 4500 base pairs, even though the longest read was substantially longer (12079 bp), over 99% of the alignments fall into this range. The MinION platform generated predominantly shorter reads, than the Sequel platform. The non-cap selected cDNA library was selected for fragments longer than 500 bp. Cap, Cap-selected cDNA sequencing on the MinION platform; Non-cap, Non-cap selected cDNA sequencing on the MinION platform; dRNA, direct RNA sequencing on the MinION platform. Sequel, cDNA sequencing on the Sequel platform. (C) shows the depth of sequencing along the HCMV genome. Reads were aligned to the HCMV strain Towne genome (LT907985.1). Below the genome, blue rectangles denote the main repeat regions of HCMV. TRL, Terminal Repeat Long; OriLyt, lytic origin of replication; IRL, Internal Repeat Long; IRS, internal Repeat Short; TRS, Terminal Repeat Short. Blue arrows mark the CDS sequences of HCMV genes. The coverage is shown on a logarithmic scale for each sequencing method. Cap, Cap-selected cDNA sequencing on the MinION platform; Non-cap, Non-cap selected cDNA sequencing on the MinION platform; dRNA, direct RNA sequencing on the MinION platform. Sequel: cDNA sequencing on the Sequel platform. All methods included polyA-selection.