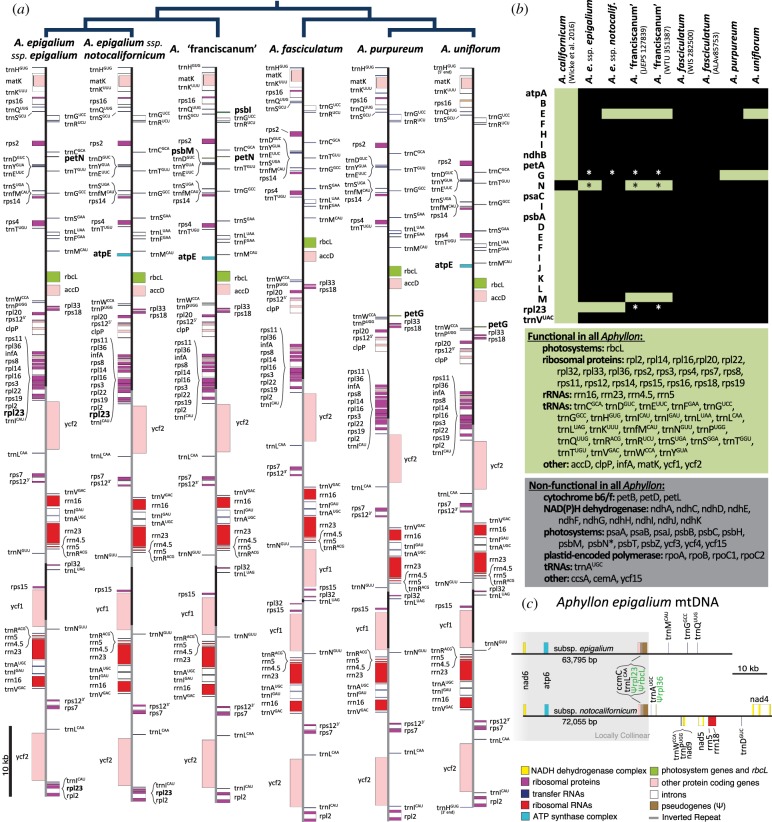
Figure 1.
(a) Annotated plastomes of each species of Aphyllon sect. Aphyllon, with their phylogenetic relationships indicated above. Only one representative of A. fasciculatum (WIS282500) and A. ‘franciscanum’ (WTU 351387) is shown. Genes not shared by all samples of Aphyllon sect. Aphyllon are labelled in bold. Location of the inverted repeats are indicated by a grey backbone. (b) Differences in plastome gene content between A. californicum and eight samples of Aphyllon sect. Aphyllon. Green cells or background indicate putatively functional genes with intact ORFs; black cells or grey background indicate that the gene is lost or probably nonfunctional. An asterisk (*) indicates ambiguities in some plastomes: petG, start codon is ATT, but the ORF is otherwise intact; petN, high amino acid divergence; psbN, intact ORF in A. fasciculatum, but high sequence divergence; rpl23, no apparent stop codon, see the electronic supplementary material, appendix D for details. (c) Mitochondrial genome fragments of A. epigalium subsp. epigalium and subsp. notocalifornicum show several pseudogenes of likely host-plastid origin (Ψ, labelled in green). Grey box indicates greater than or equal to 36642 bp locally collinear block inferred using Mauve [30].