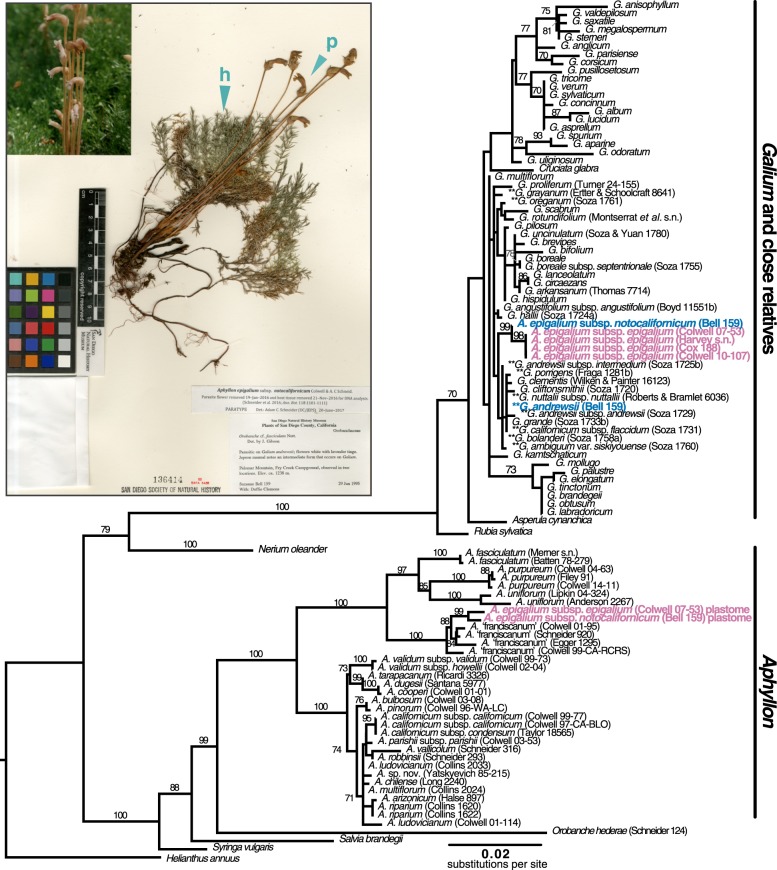
Figure 2.
Maximum-likelihood rbcL gene tree of Galium and Aphyllon homologs. Bootstrap support of greater than or equal to 70% is indicated along branches. Paralogues sequenced from A. epigalium (with pink labels) form two strongly supported clades. One is nested within a clade of Galium sequences and near a subclade containing most reported host species (denoted by **), and the other is resolved within sequences of Aphyllon sect. Aphyllon, congruent with accepted infrageneric relationships [11]. Tip labels with collector names in parentheses represent sequences generated for this study; other sequences were downloaded from GenBank. Inset: collection of A. epigalium subsp. notocalifornicum parasitizing the root of its host, Galium andrewsii (Bell and Clemons 159 [SD 136414]). Blue wedges indicate approximate locations that host (h) and parasite (p) tissue were sampled from the herbarium specimen, from which differing Galium and Galium-like rbcL genes were sequenced (blue tip labels in phylogeny). Image courtesy of San Diego Natural History Museum and used with permission.