FIGURE 4.
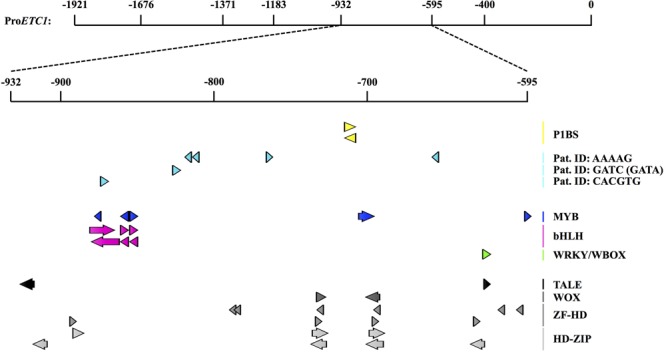
Cis-regulatory elements and motifs related to Pi responsiveness and patterning with the selected ProETC1 fragment. Detailed analysis of cis-regulatory motives in the ProETC1-932- -595 fragment using PlantPAN 2.0 (Chow et al., 2016) and manual analysis with motifs from a recent publication (Salazar-Henao et al., 2016). The figure was constructed using the CLC DNA Workbench (see text footnote 4). P1BS was previously identified by PLACE (Higo et al., 1999) mutated in this study and did not show to be causative. Recently, 14 root specific Pi-responsive motifs were identified (Salazar-Henao et al., 2016). Three of these were found in the selected ProETC1 fragment (labeled “Pat. ID”). Below, patterning related motifs extracted from a PlantPAN 2.0 analysis are shown. Orientation of motifs: +: arrows point to the right, – strand: arrow points to the left. P1BS, PHR1-binding site; Pat. ID, pattern identifier from Salazar-Henao et al. (2016); TALE, three amino acid loop extension (of homeodomains); WOX, WUS homeobox-containing; ZF-HD, zinc-finger-homeodomain; HD-ZIP, homeodomain-leucine zipper. All results and precise positions can be found in Supplementary Table S11.
