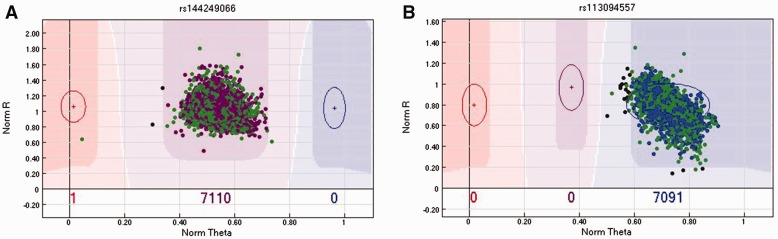
Figure 11.
(A) The first example is for SNP rs144249066 in the MEGAEX array. First, all subjects were called heterozygous [A/T], which strongly violates the HWE assumption. The HWE test had P<10−8 for this SNP in Caucasians, which means this SNP could be potentially filtered out by the HWE test. In 1000G data, this SNP was inferred as homozygous [A/A] for all Caucasians. Possible explanations are (1) the probe sequences were designed wrong or (2) they mapped to highly homologous regions. (B) The second example is for SNP rs113094557 on the MEGAEX array. This SNP does not violate HWE, and the genotype type call is [G/G] for all Caucasians; however, the genotype call for all Caucasians in 1000G is [A/A]. The SNP has two probes designed to capture alleles A and G. As the two alleles are not reverse complementary, this could not be caused by a strand issue. The only plausible explanation is that the two alleles were switched or mislabeled by Illumina during design.