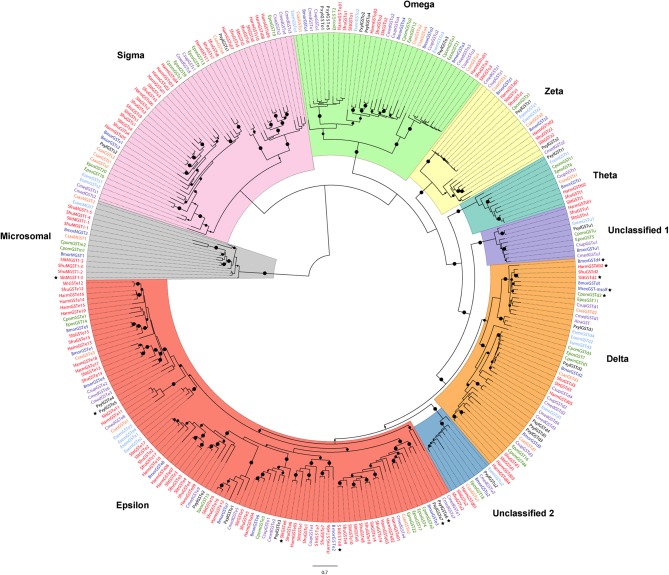
Figure 1.
Maximum-likelihood phylogeny of antennal-expressed lepidopteran GSTs. The tree was built from amino-acid sequences of GST repertoires of S. littoralis, S. frugiperda, H. armigera (Noctuidae family, branches colored in red), M. sexta, B. mori (Bombycoidea, dark blue), C. pomonella, E. postvittana (Tortricoidea; green), C. medinalis, C. suppressalis, A. transitella (Pyraloidea, purple), C. sasakii (Carposinidae, orange), P. xylostella (Plutellidae, black), and E. semipurpurella (Eriocraniidae, light blue). The MAPEG clade was used as an outgroup. Circles represent nodes highly supported by the likelihood-ratio test (small dots: aLRT > 0.9, middle dots: aLRT > 0.95, big dots: aLRT = 1). The black stars indicate antennal-enriched or antennal-specific GSTs. The scale bar represents 0.7 expected amino-acid substitutions per site.