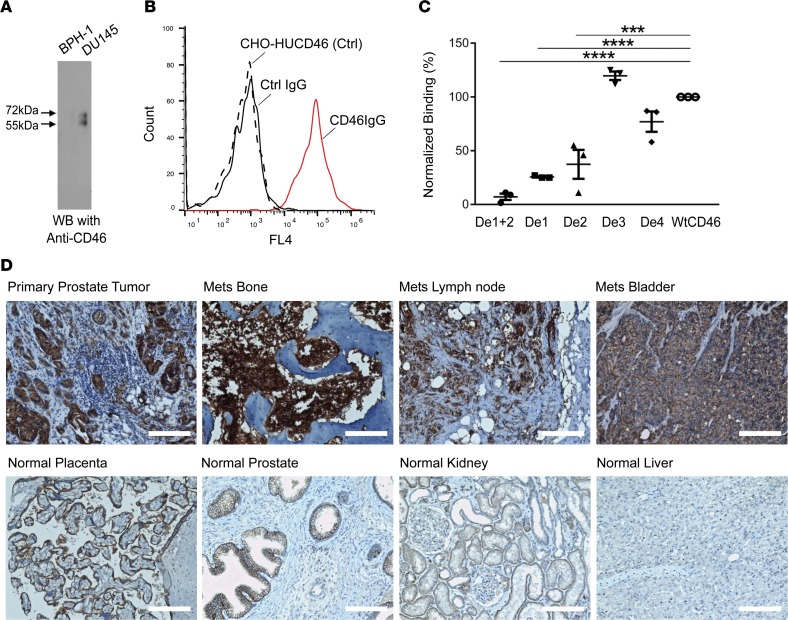
Figure 1. Identification of human CD46 as a target for prostate cancer.
(A) Verification of CD46 as the target antigen by IP and Western blot analysis using validated anti–human CD46 antibody. Two bands characteristic of human CD46 were seen in IP products from tumor (DU145) but not control nontumorigenic cells (BPH-1). CD46 is known to migrate on SDS-PAGE as 2 bands (27). (B) Verification of target identification by antibody binding to cells ectopically expressing human CD46. CHO-HUCD46: control (Ctrl), CHO-K1 cells transiently transfected with human CD46 expression plasmid without antibody incubation. Ctrl IgG: a nonbinding antibody randomly picked from naive library was incubated with CHO-HUCD46. CD46IgG: UA20 IgG was incubated with CHO-HUCD46 cellsn. (C) Epitope mapping by ectopic expression of CD46 deletion constructions followed by FACS analysis. De1+2, CD46 with Sushi domains 1 and 2 deleted. De1, domain 1 deleted. De2, domain 2 deleted. De3, domain 3 deleted. De4, domain 4 deleted. WtCD46: full-length WT CD46 that was used for normalization of FACS binding data. ***P < 0.001 (P = 0.0002), ****P < 0.0001. One-way ANOVA, Bonferroni’s multiple comparisons test. The experiment was done in triplicate. (D) IHC study of formalin fixed, paraffin-embedded prostate cancer tissues and a normal human tissue array. Top row: primary tumor and mCRPC samples with strong positive staining signals. H score for primary tumor, 211; bone mets (Mets), 295; lymph node mets 202; and bladder mets 276. Bottom row: normal tissues staining. Placental trophoblasts showed positive signals, along with prostate epithelium. Weak staining was seen for kidney and liver. H score for placenta, 167; prostate epithelium, 142; kidney, 52; and liver 12. Scale bars: 150 μm.