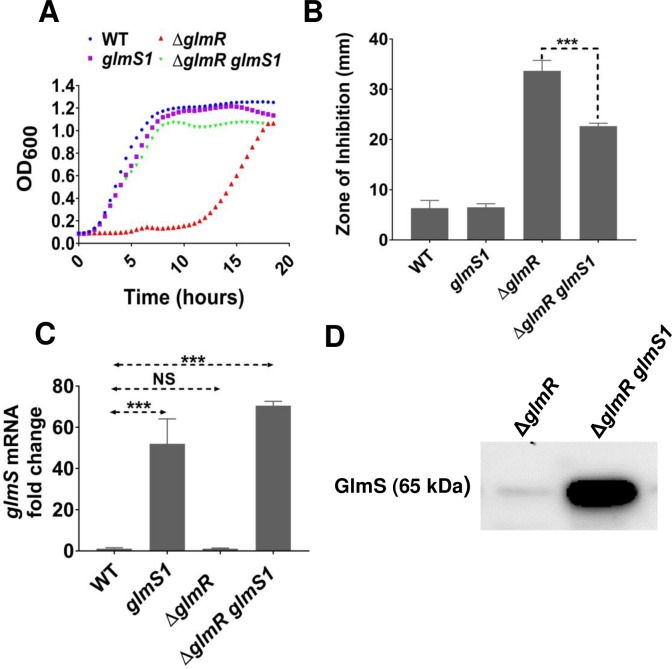
Fig 4. glmS1 suppresses ΔglmR by abolishing negative feedback regulation of glmS expression.
(A) Representative growth curves in MH medium (n>3) showing the effect of point mutation glmS1 in ΔglmR compared to WT, glmS1 and ΔglmR. (B) Disc diffusion assay showing the effect of glmS1 on CEF sensitivity phenotype of ΔglmR. 6 μg CEF was used in the assay. Standard deviation (error bars) is based on at least three biological replicates. Three asterisks indicate significant difference with P <0.001 using Tukey test. (C) qRT-PCR results showing glmS mRNA level in glmS1, ΔglmR and ΔglmR glmS1 relative to WT from cells harvested at OD600 of 0.5 grown in LB. Standard deviation (error bars) is based on at least three biological replicates. Statistical significance is determined by Tukey test where three asterisks indicate P <0.001 and NS is non-significant (P >0.05). (D) Western blot analysis of GlmS protein in ΔglmR and ΔglmR glmS1 using anti-GlmS antibodies. 5 μg of total protein was loaded in each lane.