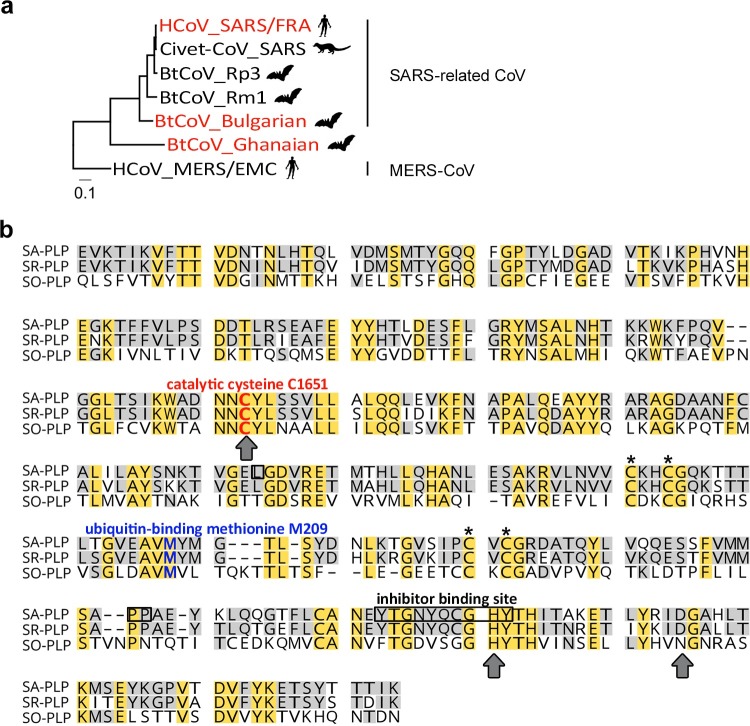
Fig 1. Phylogenetic and sequence-based analysis of the SARS-related bat coronavirus papain-like protease (SR-PLP).
(a) Phylogeny of SARS-related beta-CoVs in the PLP gene (981 bp fragment) within the nonstructural protein 3. PLP genes characterized in the study are colored in red. The right-hand column shows the species classification of the included virus clades according to the International Committee on Taxonomy of Viruses (ICTV). Phylogenetic trees of SARS-related betacoronaviruses (CoVs) were calculated by the Neighbor Joining algorithm in Geneious under the assumption of a Tamura-Nei genetic distance model. Symbols correspond to the respective host species (human, civet and bat). The scale bar refers to the genetic distance. The SARS-outlier CoV (SO-CoV) was identified in a Ghanaian Hipposideros bat. SO-CoV belongs to a novel unclassified beta-CoV species. HCoV: human CoV, FRA: SARS Frankfurt strain, BtCoV: bat CoV. The accession numbers are as follows: HCoV_SARS/FRA: AY310120, Civet CoV_SARS: AY572034, BtCoV_Rp3: DQ071615, BtCoV_Rm1: DQ022305, BtCoV_Bulgarian: GU190215, BtCoV_Ganaian: MG916963, HCoV_MERS/EMC: JX869059. (b) Amino acid sequence alignment for the comparison of SR-PLP to SA-PLP. The alignment is based on the amino acid codes by the Blosum62 algorithm in the Geneious 6 software package. The SO-CoV derived PLP (SO-PLP) was included as an outlier PLP. Yellow boxes indicate conserved residues in all sequences. The boxes in light grey indicate conserved residues in only two sequences. Residues that form the catalytic center are indicated by grey arrows below the sequences. The catalytic cysteine, which was mutated to alanine in the course of this study, is highlighted in red. The ubiquitin-binding methionine at amino acid position 209, which was mutated to arginine (M209R) in this study, is marked in blue. Zinc-binding residues, important for the three dimensional PLP structure, are indicated by asterisks above the sequences. C1651 numeration refers to the position in the SARS-CoV pp1a already used before [46]. Residues framed in black indicate the binding sites of the inhibitor compound 3e, which was used in the course of this study. SA: SARS; SR: Bulgarian; SO-PLP: Ghanaian.