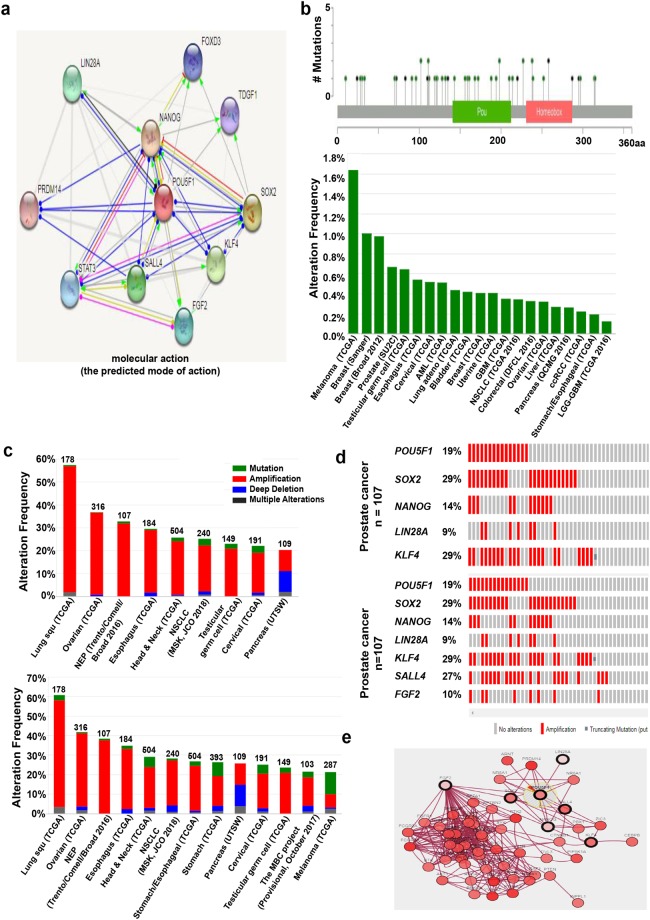
Figure 3.
Mutation and alteration frequency patterns of OCT4 (POU5F1) and its associated genes in various cancers: (a) Functional protein partner of OCT4 was predicted by STRING. Line indicates the predicted mode of molecular action. (b) Mutation diagram of POU5F1 in different cancer types across protein domains. POU5F1 mutation frequencies are the highest in melanoma and POU5F1 mutation mere more frequent in N-domain than in the C-domain. (c) The alteration frequency of a five-gene signature (POU5F1, SOX2, NANOG, LIN2BA, and KLF4) was determined using cBioPortal and is shown on the top. The alteration frequency of a seven-gene signature (POU5F1, SOX2, NANOG, LIN2BA, KLF4, SALL4, and FGF2) was determined using cBioPortal and is shown on the bottom. Only cancer types containing >100 samples and an alteration frequency of >20% are shown. The alteration frequency included deletions (blue), amplification (red), multiple alterations (grey), or mutation (green). The total number of samples for each cancer type is indicated by the numbers at the top of each column. Prostate cancer types frequently amplify POU5F1. We used the Oncoprint feature of cBioPortal to determine the copy number alteration frequency of each gene in POU5F1 within selected cancer subtypes. (d) The percentages of alterations in five genes and seven genes in the prostate cancer. Grey bars along a vertical line represent the same sample evaluated for amplification (red), deep deletion (blue), missense mutation (green), truncating mutation (black), or in-frame mutation (brown). (e) The interactions between POU5F1 and its associated gene alterations were searched in cBio Cancer Genomics Portal. Network view of the POU5F1 neighborhood in prostate cancer. Darker red indicates increased frequency of alteration (defined by mutation, copy number amplification, or homozygous deletion) in prostate cancer.