Abstract
Forkhead box (FOX)K2 (FOXK2) is a member of the FOX transcription factor family. It has been suggested previously that FOXK2 is required to suppress tumor growth; however, the exact role of FOXK2 in gastric cancer remains to be elucidated. In the present study, the association between FOXK2 expression and the clinicopathological characteristics of patients with gastric cancer was investigated. The prognostic value of FOXK2 expression and the significance of clinicopathological parameters in the overall survival (OS) and progression-free survival of patients were also determined by survival analysis. To investigate the functional roles of FOXK2, it was downregulated in BGC-823 cells using small interfering (si)RNA, and upregulated using a FOXK2 plasmid. Colony formation, Cell Counting Kit-8 and cell proliferation analyses were conducted to examine the proliferation of gastric cancer cells. Transwell and wound-healing assays were performed to investigate the effect of FOXK2 expression on gastric cancer cell migration and invasion. The clinical data demonstrated that FOXK2 expression was reduced in high-grade gastric cancer tissues, and a low level of FOXK2 expression indicated a poor prognosis. The data obtained from the Human Protein Atlas revealed that patients with gastric cancer and a high level of FOXK2 expression had a longer OS time. The results of colony formation assays, Transwell and wound healing assays demonstrated that FOXK2 repressed the proliferation, invasion and migration of gastric cancer cells, respectively. The findings indicated that FOXK2 may serve as a promising therapeutic target in gastric cancer. Taken together, the findings of the present study demonstrated that FOXK2 functions as a tumor suppressor in gastric cancer; the loss of FOXK2 may induce the growth and invasion of gastric cancer cells.
Keywords: forkhead box K2, gastric cancer, epithelial-mesenchymal transition, proliferation, invasion
Introduction
Gastric cancer is the third leading cause of cancer-associated mortality worldwide (1–3). Although the diagnosis and treatment of gastric cancer has improved over the past few decades, new cases and estimated mortalities are increasing every year (4). The development of gastric cancer is a complex process. Several studies have investigated the mechanism of gastric cancer and numerous therapeutic targets have been explored; however, effective therapeutic targets have not yet been identified (3,5). Aberrant activation of epithelial-mesenchymal transition (EMT) is a crucial process in gastric carcinogenesis (5,6). Typically, levels of E-cadherin, N-cadherin, twist family BHLH transcription factor 1 (TWIST) and snail family transcriptional repressor 1 (SNAIL) are detected to evaluate the EMT process. Forkhead box (FOX)K2 is a member of the FOX transcription factor family; FOXK2 is a vital protein that is phosphorylated by the cyclin-dependent kinase (CDK) complex (7). FOXK2 regulates numerous genes involved in cell adhesion, motility, metabolism, apoptosis and tumorigenesis (8,9). Previous research has demonstrated that the overexpression of FOXK2 suppresses EMT in non-small cell lung cancer, through inhibition of N-cadherin and β-catenin expression (10).
Gene expression is regulated by transcription and relies on transcription factors to enable or disable gene expression. It has been reported that FOXK2 recruits distinct corepressor complexes, including proteins such as SIN3 transcription regulator family member A, RE1 silencing transcription factor, nuclear receptor corepressor 2 and histone deacetylases, which are important for gene transcription (11,12). A recent report demonstrated that FOXK2 interacts with polycomb complex molecules and recruits tumor suppressor proteins to modify the structure of chromatin (13).
FOXK2 is involved in several signaling pathways, including the mechanistic target of rapamycin, Wnt and AKT serine/threonine kinase 1 pathways. These signaling pathways regulate cell proliferation and death by affecting the expression of important genes via the transcriptional repressor FOXK2 (10,14). FOXK2 not only affects tumorigenesis, but also tumor drug resistance. A previous study demonstrated that FOXK2 improves MCF-7 cell susceptibility to paclitaxel by regulating cyclin B1 (12). However, the function and mechanism of FOXK2 in gastric cancer remains largely unknown. In the present study, the function of FOXK2 in the development and progression of gastric cancer was investigated.
Materials and methods
Patients and tissue specimens
The present study was approved by the Ethics Committee of Tianjin Nankai Hospital (Tianjin, China). Clinical data were collected between July 2016 and July 2017, and a total of 150 patients were recruited to the study from Tianjin Nankai Hospital. Written informed consent was obtained from all patients. Clinical information, including age, sex, differentiation grade and tumor size, was obtained (Table I). Tissue samples of gastric cancer and para-tumor tissue were excised from the patients with gastric cancer during resection. Tissue samples were fixed immediately in 4% neutral-buffered formalin at room temperature for 24 h and subsequently processed to prepare paraffin-embedded sections (1×1×0.5 cm).
Table I.
Association between FOXK2 expression and gastric cancer clinicopathological features.
| FOXK2 expression | ||||
|---|---|---|---|---|
| Clinicopathological feature | Patients (n) | Low (n) | High (n) | P-value |
| Age (years) | 0.705 | |||
| <50 | 86 | 43 | 43 | |
| ≥50 | 64 | 34 | 30 | |
| Sex | 0.142 | |||
| Male | 75 | 34 | 41 | |
| Female | 75 | 43 | 32 | |
| Tumor size (cm) | 0.002 | |||
| <5 | 77 | 30 | 47 | |
| ≥5 | 73 | 47 | 26 | |
| Differentiation | 0.042 | |||
| Well | 22 | 9 | 13 | |
| Moderate | 102 | 49 | 53 | |
| Poor | 26 | 19 | 7 | |
FOXK2, forkhead box K2.
FOXK2 expression analysis in datasets
FOXK2 expression data in gastric cancer were collated from the Human Protein Atlas (15).
Immunohistochemical staining
Among the samples, 22 tumors were classified as well differentiated, 102 as moderately differentiated and 26 as poorly differentiated. Formalin-fixed tissue samples were prepared prior to immunohistochemical staining. Samples were stained using the avidin-biotin complex method. Primary antibodies specific for FOXK2 (1:500; cat. no. ab50946; Abcam, Cambridge, MA, USA) were incubated with sections overnight at 4°C. The samples were then incubated with a biotin-conjugated goat anti-rabbit immunoglobulin G secondary antibody (1:100; cat. no. TA130016; OriGene Technologies, Inc., Beijing, China) at 37°C for 1 h. The expression of FOXK2 was detected by coloration with DAB, and the procedure was performed as previously described (16). Staining intensity was scored as follows with an inverted microscope (Olympus Corporation, Tokyo, Japan): 0, negative; 1, weakly positive; 2, moderately positive and 3, strongly positive. The percentage of FOXK2-positive cells was therefore scored as 0 (0%), 1 (1–25%), 2 (26–50%) or 3 (>50%).
Cell culture, chemical reagents and antibodies
The human gastric cancer cell line BGC-823 was obtained from the China Academia Sinica Cell Repository (Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) under 5% CO2 at 37°C. FOXK2 (cat. no. ab50946; Abcam), cleaved caspase-3 (cat. no. ab2302; Abcam), E-cadherin (cat. no. ab15148; Abcam) and N-cadherin (cat. no. ab18203; Abcam) antibodies were purchased from Abcam. GAPDH antibody (cat. no. TA802519) was purchased from OriGene Technologies, Inc.
Transfection
For transient cell transfection, gastric cancer cells (1.5×106 cells/well) were seeded in 6-well plates and cultured overnight, and subsequently transfected with FOXK2 plasmids encoding human FOXK2 or empty vector. The FOXK2 overexpression plasmid (plasmid no. S57120) and empty vector were purchased from GenScript (Nanjing, China). FOXK2 small interfering (si)RNA (si-FOXK2; 50 nM; sense, 5′-GAGTTCGAGTATCTGATGA-3 and antisense, 5′-GCGAACACGTACACTGTCT-3′) and negative control siRNA (50 nM; cat. no. siN05815122147-1-5) were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China) and transfected with X-tremeGENE™ 9 DNA transfection reagent (cat. no. 06365787001; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and X-tremeGENE™ siRNA transfection reagent (cat. no. 04476093001; Sigma-Aldrich; Merck KGaA). Plasmid transfection (1 µg/well) was performed with Lipofectamine® 3000 (cat. no. L300001; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells were collected and seeded for assays 48 h post-transfection.
Western blot analysis
Antibodies against FOXK2, cleaved caspase-3, N-cadherin, E-cadherin and GAPDH were used for western blot analysis, as described previously (14). The cells were washed with PBS three times and protein was extracted in radioimmunoprecipitation assay lysis buffer (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) with 1% phenylmethanesulfonyl fluoride. Protein concentration was determined with a bicinchoninic acid protein assay. Proteins (40 µg/lane) were separated by 10% SDS-PAGE. The separated proteins were transferred to polyvinylidene fluoride membranes and blocked in 5% bovine serum albumin (Beijing Solarbio Science & Technology Co., Ltd.) at 37°C for 1 h. Following this, membranes were incubated with primary antibodies against FOXK2, cleaved caspase-3, N-cadherin, E-cadherin and GAPDH (1:1,000 dilution) at 4°C for 12 h, and subsequent incubation with horseradish peroxidase-conjugated secondary antibody (1:2,000 dilution; cat. no. ZDR-5306; OriGene Technologies, Inc., Beijing, China) at room temperature for 1 h. Protein expression was visualized using with an enhanced chemiluminescence kit (cat. no. WBKLS0500; Merck KGaA). Bands were analyzed with ImageJ software (version 1.51j8; National Institutes of Health, Bethesda, MD, USA).
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Equal amounts of RNA were converted into cDNA with a PrimeScript RT reagent kit (Promega Corporation, Madison, WI, USA) according to manufacturer's protocol. Relative expression levels of FOXK2 were determined by PCR with a GoTaq® Real-Time PCR system (Promega Corporation). The primer sequences for FOXK2 were as follows: Forward, 5′-AAGAACGGGGTATTCGTGGAC-3′ and reverse, 5′-CTCGGGAACCTGAATGTGC-3′. The reference gene was GAPDH, and the primer sequences for GAPDH were as follows: Forward, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse, 5′-GCCATCACGCCACAGTTTC-3′. The PCR conditions were as follows: 30 cycles of 94°C for 30 sec, 56°C for 30 sec, 72°C for 90 sec, and a final extension at 72°C for 5 min. Quantification was performed using the 2−∆∆Cq method (17).
Colony formation and Cell Counting Kit (CCK)-8 assays. Tumor cells transfected with si-FOXK2 or FOXK2 plasmid were cultured at 2,000 cells/well in 6-well plates. The cells were allowed to grow for 14 days, and the medium was changed every 3 days. Colonies were subsequently fixed with 4% paraformaldehyde for 1 h at room temperature, stained with 0.5% crystal violet for 10 min at room temperature and counted under an inverted microscope (Olympus Corporation).
For assessment of cell proliferation, a CCK-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was performed according to the manufacturer's protocol. To determine the effect of FOXK2 on cell proliferation, tumor cells were transfected with si-FOXK2 or FOXK2 plasmid, seeded into a 96-well plate at a density of 2×103 cells in 100 µl culture medium containing 10% FBS and cultured overnight. All experiments were performed in triplicate. The medium was subsequently replaced with 100 µl fresh medium containing 10% CCK-8 reagent, and cells were incubated for 3.5 h at 37°C. At 0, 24, 48, 72, 96 and 120 h, absorbance was measured at 450 nm using an ELx800 microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
To determine the effect of FOXK2 on cell proliferation, cells were also assessed with a colony formation assay. Tumor cells transfected with si-FOXK2 or FOXK2 plasmid were seeded into each well of a 12-well plate at a density of 5×104 cells in 2 ml culture medium containing 10% FBS, and were cultured overnight. All experiments were performed in triplicate. The cells were incubated at 37°C. At 0, 24, 48, 72, 96 and 120 h, the cells were counted following trypsin enzyme digestion.
Apoptosis assay
Annexin V/propidium iodide (PI) staining was performed to quantify cell apoptosis. Transfected cells (8×106) in the logarithmic growth phase were collected and subjected to Annexin V/PI staining using an Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection kit (BioVision, Inc., Milpitas, CA, USA) according to the manufacturer's protocol. The resulting fluorescence was measured by flow cytometry using a FACS flow cytometer (BD Biosciences, San Jose, CA, USA). Obtained data were analyzed via BD Cell Quest Pro™ software (version 5.1; BD Biosciences).
Transwell invasion assay
The ability of cells to invade was assessed using Matrigel-coated Transwell membranes (BD Biosciences). After 30 min incubation at 37°C, the Matrigel solidified and served as an extracellular matrix for tumor cell invasion analysis. The upper chamber contained ~5×104 cells in 200 µl DMEM without serum. The lower chamber contained DMEM with 10% FBS. The cells were then incubated for 48 h at 37°C in an atmosphere containing 5% CO2. Cell invasion to the underside of the Matrigel-coated membrane was subsequently fixed with 4% paraformaldehyde for 10 min at room temperature, stained with 0.5% crystal violet for 10 min at room temperature followed by imaging and counting under an inverted microscope at ×200 magnification (Olympus Corporation). The results are expressed as the average number of invasive cells per field.
Wound-healing assay
BGC-823 cells were transfected with si-FOXK2 or FOXK2 plasmid at 37°C for 48 h, seeded into 6-well plates (5×106 cells/well), cultured in serum-free medium and a straight wound was created using a pipette tip. Cells were further incubated at 37°C in an atmosphere containing 5% CO2 for 24 h. Images of wound healing were captured under an inverted microscope at ×100 magnification (Olympus Corporation, Tokyo, Japan). The percentage of wound closure was the rate of migration distance compare to the control group. Results were analyzed with ImageJ software (version 1.51j8; National Institutes of Health).
Statistical analysis
Data are presented as the means ± standard deviation of three independent experiments. Statistical analyses were performed in SPSS version 16.0 (SPSS, Inc., Chicago, IL, USA). Two groups were compared using Student's t-test, whereas multiple groups were compared using one-way analysis of variance followed by the Student-Newman-Keuls method. Survival curves were plotted using the Kaplan-Meier method, and the differences between the survival curves were examined by the log-rank test. Cox proportional hazards models were used to identify factors with an independent influence on survival (18–20). P<0.05 was considered to indicate a statistically significant difference.
Results
FOXK2 is downregulated in gastric cancer, and high FOXK2 expression indicates a good prognosis
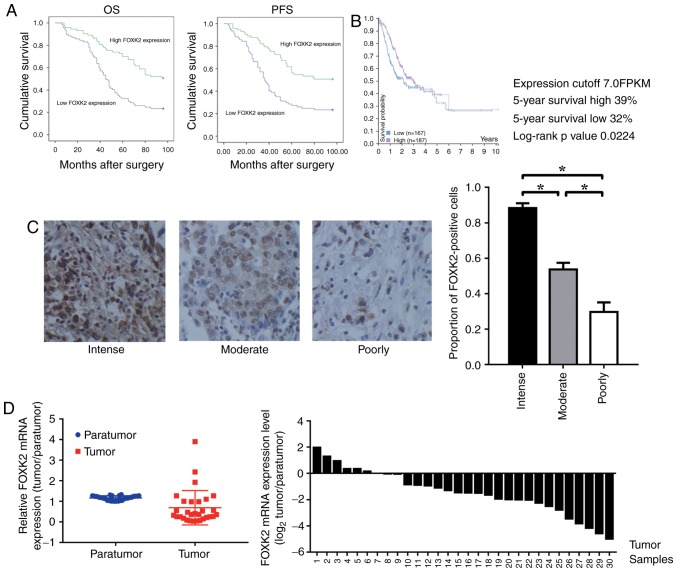
A total of 150 patients were examined (male:female, 1:1), and the associations between FOXK2 expression and clinicopathological characteristics were investigated. Among the samples, 22 tumors were well differentiated, 102 cases were moderately differentiated and 26 were poorly differentiated. It was demonstrated that the expression of FOXK2 was positively correlated with tumor differentiation (P<0.05; Table I). There was no significant association between FOXK2 expression with sex and age (P>0.05; Table I). Cox regression analyses revealed a significant association between overall survival and tumor size, FOXK2 expression (P<0.001; Table II). The prognostic value of FOXK2 expression with regards to the OS and PFS of patients was determined by survival analysis. The results revealed that high FOXK2 expression was associated with an improved prognosis (Fig. 1A).
Table II.
Univariate and multivariate analyses of prognostic parameters in patients with gastric cancer in terms of overall survival.
| Parameter | Univariate log-rank test (P-value) | Cox multivariate analysis (P-value) | Risk |
|---|---|---|---|
| Age (<50 vs. ≥50 years) | 0.153 | – | – |
| Sex (male vs. female) | 0.095 | – | – |
| Differentiation (well, moderate, poor) | – | <0.001 | 2.637 |
| Tumor size (<4 vs. ≥4 cm) | <0.001 | 0.008 | 1.840 |
| Forkhead box K2 expression (low vs. high) | <0.001 | 0.006 | 0.545 |
Figure 1.
Expression of FOXK2 in gastric cancer tissues. (A) Kaplan-Meier analysis of OS and PFS based on FOXK2 expression in patients with gastric cancer (log-rank test, P<0.01). (B) Kaplan-Meier analysis for survival was performed with data obtained from The Human Protein Atlas (log-rank test, P=0.0224). (C) Immunohistochemical staining of FOXK2 in various gastric cancer grades (magnification, ×200). The results were analyzed with one-way analysis of variance followed by Student-Newman-Keuls method; *P<0.05. (D) Compared with para-tumor tissue samples, relative FOXK2 mRNA expression was significantly lower in tumor tissue samples. FOXK2, forkhead box K2; OS, overall survival; PFS, progression free survival.
Kaplan-Meier survival analysis was performed to analyze the association between FOXK2 mRNA expression and patient survival. Data on gastric cancer were obtained from the Pathology Atlas in The Human Protein Atlas (http://www.proteinatlas.org/). The results revealed that a high level of FOXK2 expression was associated with improved prognosis (P=0.0224; Fig. 1B).
FOXK2 expression was reduced in patients with high-grade gastric cancer. The proportion of FOXK2-positive cells in the intense, moderately and poorly differentiated gastric cancer tissues was 88.3±4.7, 53.7±6.5 and 29.7±9.5%, respectively (P<0.05; Fig. 1C). Compared with the para-tumor tissue, the relative mRNA expression levels of FOXK2 were significantly lower in the tumor tissue (Fig. 1D).
FOXK2 regulates gastric cancer cell apoptosis
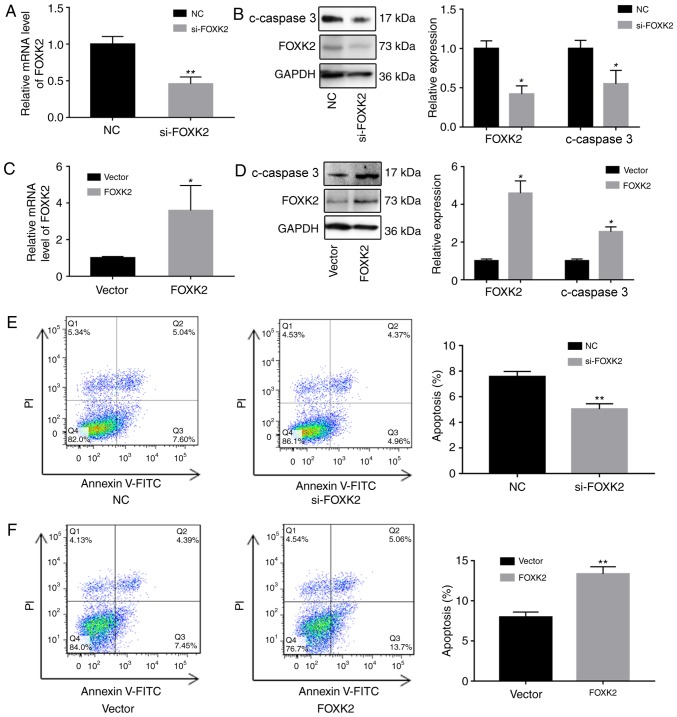
To investigate the functions of FOXK2, FOXK2 was downregulated in BGC-823 cells using siRNA, and FOXK2 was upregulated using a FOXK2 plasmid. Alterations in the expression of FOXK2 were verified by RT-qPCR and western blotting. Western blot analysis revealed that increased FOXK2 expression reduced the upregulation of cleaved caspase-3 (Fig. 2A-D).
Figure 2.
FOXK2 expression affects c-caspase-3 expression and BGC-823 cell apoptosis. (A) si-FOXK2 transfection effectively reduced FOXK2 mRNA expression in BGC-823 cells, as well as (B) FOXK2 and c-caspase-3 protein expression. (C) FOXK2 overexpression plasmid transfection successfully increased FOXK2 mRNA. (D) FOXK2 and c-caspase-3 protein expression also increased. (E) si-FOXK2 decreased BGC-823 cell apoptosis, whereas (F) FOXK2 overexpression increased BGC-823 cell apoptosis. *P<0.05, **P<0.01 vs. the corresponding control group. c-, cleaved; FITC, fluorescein isothiocyanate; FOXK2, forkhead box K2; NC, negative control; PI, propidium iodide; si-, small interfering RNA.
The effects of FOXK2 on apoptosis were subsequently determined by western blot analysis and flow cytometry. The percentage of apoptotic cells in the si-FOXK2 group (5.03±0.44%) was significantly reduced compared with in the negative control (NC) group (7.56±0.34%; P<0.01; Fig. 2E). Furthermore, the percentage of apoptotic cells in the FOXK2 group (13.4±0.89%) was significantly increased compared with in the vector group (7.9±0.64%; P<0.01; Fig. 2F). Cells in the lower right quadrant were early apoptotic cells, and the upper right quadrant was designated as late apoptotic cells. These results indicated that upregulation of FOXK2 expression induced early apoptosis, whereas FOXK2 expression in gastric cancer cells had no effect on late apoptosis.
FOXK2 inhibits gastric cancer cell invasion
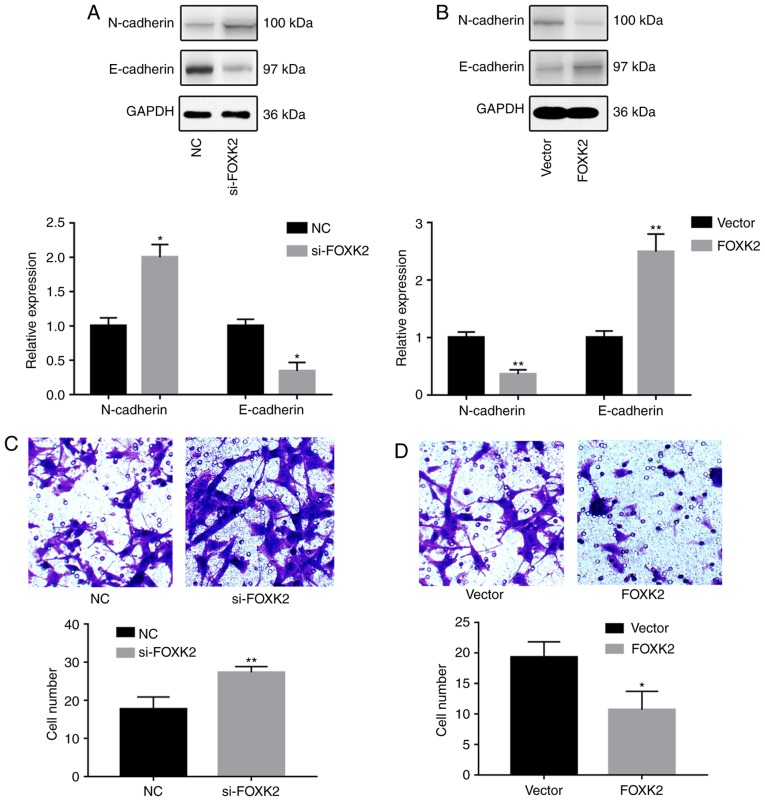
To investigate the effects of FOXK2 on gastric cancer cell migration and invasion, western blotting, Transwell and wound-healing assays were conducted. Western blot analysis revealed that FOXK2 knockdown increased the expression of N-cadherin and decreased the expression of E-cadherin (P<0.05; Fig. 3A). Conversely, FOXK2 upregulation decreased the expression of N-cadherin and increased the expression of E-cadherin (P<0.01; Fig. 3B). Transwell assays were conducted in order to investigate the effects of FOXK2 on gastric cancer invasion. The results revealed that the invasion rate was increased by 54.7% in BGC-823 cells transfected with si-FOXK2 (P<0.01; Fig. 3C). The invasion rate was decreased by 44.8% following FOXK2 plasmid transfection (P<0.05; Fig. 3D).
Figure 3.
FOXK2 regulates gastric cancer cell invasion. Western blot analysis demonstrated alterations in N-cadherin and E-cadherin expression following (A) si-FOXK2 or (B) FOXK2 plasmid transfection. Transwell assays were performed to assess the invasion of cells transfected with (C) si-FOXK2 or (D) FOXK2 plasmid. Magnification, ×200. *P<0.05, **P<0.01 vs. the corresponding control group. FOXK2, forkhead box K2; NC, negative control; si-, small interfering RNA.
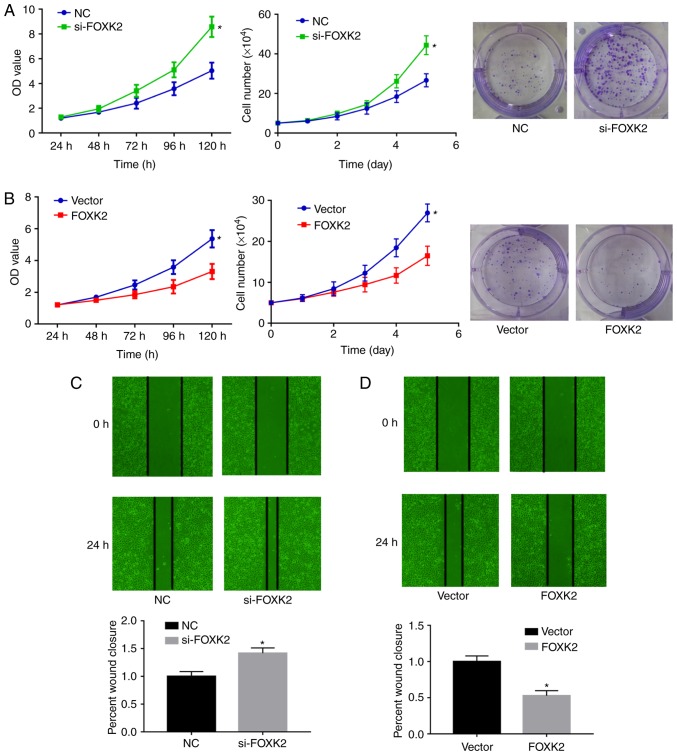
FOXK2 regulates gastric cancer cell proliferation and migration. To clarify the role of FOXK2 in gastric cancer, colony formation, CCK-8 and cell proliferation assays were performed to examine BGC-823 cell proliferation. The expression levels of FOXK2 in BGC-823 gastric cancer cells were downregulated by si-FOXK2 and upregulated by FOXK2 plasmid transfection. Colony formation, CCK-8 and cell proliferation assays demonstrated that si-FOXK2 significantly increased the growth of gastric cancer cells compared with the NC group (P<0.05; Fig. 4A), whereas FOXK2 plasmid transfection inhibited BGC-23 cell growth compared with the vector group (P<0.05; Fig. 4B). Average colony numbers in the NC and si-FOXK2 groups were 24.7±5.7 and 46±3.2, respectively (P<0.05). Average colony numbers in the vector and FOXK2 groups were 34±7.55 and 16.7±3.1, respectively (P<0.05; Fig. 4A and B).
Figure 4.
FOXK2 regulates gastric cancer cell proliferation. (A) Growth curve of cultured BGC-823 cells and the colony formation assay demonstrated that si-FOXK2 transfection induced cell proliferation, whereas (B) FOXK2 overexpression inhibited cell proliferation. (C) Wound-healing assays demonstrated that si-FOXK2 transfection increased cell migration, whereas (D) FOXK2 plasmid transfection inhibited cell migration. Magnification, ×100. *P<0.05 vs. the corresponding control group. FOXK2, forkhead box K2; NC, negative control; si-, small interfering RNA.
Wound-healing assays were conducted to investigate the function of FOXK2 in gastric cancer cell migration. In BGC-823 cells, the migration rate was increased by 32% following si-FOXK2 transfection compared with the NC group (Fig. 4C). Migration rate was decreased by 47.3% following FOXK2 plasmid transfection (P<0.05). Taken together, these results indicated that FOXK2 upregulation inhibited the migration of gastric cancer cells (P<0.05; Fig. 4C and D).
Discussion
Gastric cancer has a poor prognosis and early diagnosis is difficult. The most effective method of early diagnosis is gastroscopy and pathological biopsy (21). Pathological diagnosis largely depends on cell morphology, and the most commonly used marker proteins are Ki-67 and B-cell lymphoma 2 (22,23). Numerous studies have been conducted to identify novel therapeutic targets, with limited success (24–26).
In the present study, FOXK2 expression was evaluated in gastric cancer tissues with different tumor grades. FOXK2 expression was downregulated in high-grade gastric cancer, compared with in low-grade tissue. The results revealed that high FOXK2 expression indicated a better prognosis, thus indicating that FOXK2 may serve as a therapeutic target in gastric cancer and as a prognostic marker for patients with gastric cancer at different stages. In addition, the results demonstrated that FOXK2 overexpression reduced cell invasion, growth and proliferation. EMT is a critical step in cancer metastasis (27,28); during EMT, epithelial cells change phenotype and obtain the characteristics of mesenchymal cells, gaining the ability to migrate and contribute to tumor metastasis (29–31). EMT is accompanied by an alteration in the expression of several proteins, including N-cadherin, E-cadherin, TWIST, SNAIL and β-catenin (16). FOXK2 has previously been reported to act as a critical mediator of EMT in certain tumors, such lung cancer (10). Notably, a study demonstrated that overexpression of FOXK2 decreased the level of N-cadherin and increased the level of E-cadherin (32), indicating that FOXK2 regulates the EMT process and may act as the core protein during EMT. In addition, a previous study revealed the relationship between FOXK2 and CDK; FOXK2 serves a crucial role in the cell cycle and is phosphorylated in a cell cycle-dependent manner to inhibit cell cycle progression (7). Furthermore, FOXK2 expression is associated with tumor development, as well as a poor prognosis and outcome (8). The CDK complex mediates the phosphorylation of FOXK2, and Ser368 and Ser423 are the two sites that control the activity of FOXK2 (7). FOXK2 regulates the phosphorylation of CDK and arrests cells at the G2/M phase (6).
The functional mechanism by which FOXK2 inhibits tumor growth remains unclear. It has been reported that FOXK2 regulates various signaling pathways, including Wnt. Furthermore, FOXK2 acts as a core protein in the function of several oncogenes. Nestal de Moraes et al (11) demonstrated that FOXK2 expression is negatively correlated with enhancer of zeste homolog 2 (EZH2) expression (10). EZH2 is expressed at a high level in several malignancies, including gastric and breast cancer (33). In addition, it has been reported that the levels of EZH2 are associated with tumor stage and prognosis (11); EZH2 is one protein through which FOXK2 functions.
In conclusion, the findings of the present study demonstrated that FOXK2 functions as a tumor suppressor in gastric cancer. In support of this approach to cancer treatment, the increasing availability of molecular diagnostic techniques may help identify patients who are more likely to respond to related drugs, such as paclitaxel (34). However, the mechanism underlying the effects of FOXK2 on gastric cancer is unclear, and the mechanism by which FOXK2 suppresses the growth of gastric cancer cells requires further research.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Chinese National Natural Science Foundation (grant no. 81671380), the Key Project of Tianjin National Natural Science Foundation (grant no. 17JCZDJC35900), the Basic and Advanced Technology Research Foundation (grant no. 152300410162) and the Science and Technology Development Foundation from the Science and Technology Department of Henan Province (grant no. 172102310103).
Availability of data and materials
The datasets generated and/or analyzed during the current study available from the corresponding author on reasonable request.
Authors' contributions
BW, HZ, XL, XW and DW designed the study. XL, XW and WN performed the data collection and wrote the manuscript. BW and HZ performed the data analysis. XL, XW and DW performed the in vitro assays. BW, HZ, XL, XW and WN critically revised and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Tianjin Nankai Hospital (Tianjin, China) and all patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lansdorpvogelaar I, Kuipers EJ. Screening for gastric cancer in Western countries. Gut. 2016;65:543–544. doi: 10.1136/gutjnl-2015-310356. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Apicella M, Corso S, Giordano S. Targeted therapies for gastric cancer: Failures and hopes from clinical trials. Oncotarget. 2017;8:57654–57669. doi: 10.18632/oncotarget.14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsuji T, Ibaragi S, Hu GF. Epithelial-mesenchymal transition and cell cooperativity in metastasis. Cancer Res. 2009;69:7135–7139. doi: 10.1158/0008-5472.CAN-09-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roussos ET, Keckesova Z, Haley JD, Epstein DM, Weinberg RA, Condeelis JS. AACR special conference on epithelial-mesenchymal transition and cancer progression and treatment. Cancer Res. 2010;70:7360–7364. doi: 10.1158/0008-5472.CAN-10-1208. [DOI] [PubMed] [Google Scholar]
- 6.Marais A, Ji Z, Child ES, Krause E, Mann DJ, Sharrocks AD. Cell Cycle-dependent regulation of the forkhead transcription factor FOXK2 by CDK·cyclin complexes. J Biol Chem. 2010;285:35728–35739. doi: 10.1074/jbc.M110.154005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian Y, Xia S, Feng Z. Sox9 mediated transcriptional activation of FOXK2 is critical for colorectal cancer cells proliferation. Biochem Biophys Res Commun. 2017;29:475–481. doi: 10.1016/j.bbrc.2016.12.119. [DOI] [PubMed] [Google Scholar]
- 8.Shi Z, Liu J, Yu X, Huang J, Shen S, Zhang Y, Han R, Ge N, Yang Y. Loss of FOXF2 expression predicts poor prognosis in hepatocellular carcinoma patients. Ann Surg Oncol. 2016;23:211–217. doi: 10.1245/s10434-015-4515-2. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Jiang S, Hu F, Xu Y, Wang T, Mei Q. Foxk2 inhibits non-small cell lung cancer epithelial-mesenchymal transition and proliferation through the repression of different key target genes. Oncol Rep. 2017;37:2335–2347. doi: 10.3892/or.2017.5461. [DOI] [PubMed] [Google Scholar]
- 10.Shan L, Zhou X, Liu X, Wang Y, Su D, Hou Y, Yu N, Yang C, Liu B, Gao J, et al. FOXK2 elicits massive transcription repression and suppresses the hypoxic response and breast cancer carcinogenesis. Cancer Cell. 2016;30:708–722. doi: 10.1016/j.ccell.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Nestal de Moraes G, Khongkow P, Gong C, Yao S, Gomes AR, Ji Z, Kandola N, Delbue D, Man EP, Khoo US, et al. Forkhead box K2 modulates epirubicin and paclitaxel sensitivity through FOXO3a in breast cancer. Oncogenesis. 2015;4:e167. doi: 10.1038/oncsis.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Ji Z, Webber A, Sharrocks AD. Genome-wide binding studies reveal DNA binding specificity mechanisms and functional interplay amongst Forkhead transcription factors. Nucleic Acids Res. 2016;44:1566–1578. doi: 10.1093/nar/gkv1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowman CJ, Ayer DE, Dynlacht BD. Foxk proteins repress the initiation of starvation-induced atrophy and autophagy programs. Nat Cell Biol. 2014;16:1202–1214. doi: 10.1038/ncb3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Z, Qian X, Li L, Zhang J, Zhu S, Zhu J, Chen L, Zhang K, Han L, Yu S, et al. Nuclear translocation of β-catenin is essential for glioma cell survival. J Neuroimmune Pharmacol. 2012;7:892–903. doi: 10.1007/s11481-012-9354-3. [DOI] [PubMed] [Google Scholar]
- 15.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 16.Choi IJ, Lee NR, Kim SG, Lee WS, Park SJ, Kim JJ, Lee JH, Kwon JW, Park SH, You JH, et al. Short-term outcomes of endoscopic submucosal dissection in patients with early gastric cancer: A prospective multicenter cohort study. Gut Liver. 2016;10:739–748. doi: 10.5009/gnl15466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Guo JG, Guo CC, He ZQ, Cai XY, Mou YG. High MMP-26 expression in glioma is correlated with poor clinical outcome of patients. Oncol Lett. 2018;16:2237–2242. doi: 10.3892/ol.2018.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aoyama T, Yoshikawa T, Fujikawa H, Hayashi T, Ogata T, Cho H, Yamada T, Hasegawa S, Tsuchida K, Yukawa N, et al. Prognostic factors in stage IB gastric cancer. World J Gastroenterol. 2014;20:6580–6585. doi: 10.3748/wjg.v20.i21.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maounis NF, Dráberová E, Trakas N, Chorti M, Riga D, Tzannis K, Kanakis M, Voralu K, Ellina E, Mahera E, et al. Expression of γ-tubulin in non-small cell lung cancer and effect on patient survival. Histol Histopathol. 2018:18027. doi: 10.14670/HH-18-027. [DOI] [PubMed] [Google Scholar]
- 21.Liu G, Xiong D, Zeng J, Chen B, Huang Z. Clinicopathological and prognostic significance of Ki-67 immunohistochemical expression in gastric cancer: A systematic review and meta-analysis. Onco Targets Ther. 2017;10:4321–4328. doi: 10.2147/OTT.S143089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min KW, Kim DH, Son BK, Kim DH, Kim EK, Seo J, Ahn SB, Jo YJ, Park YS, Ha J. A High Ki67/BCL2 index could predict lower disease-free and overall survival in intestinal-type gastric cancer. Eur Surg Res. 2017;58:158–168. doi: 10.1159/000448945. [DOI] [PubMed] [Google Scholar]
- 23.Nishizawa T, Yahagi N. Long-term outcomes of using endoscopic submucosal dissection to treat early gastric cancer. Gut Liver. 2018;12:119–124. doi: 10.5009/gnl17095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, Liu X, Gong P, Song W, Zhou M, Li Y, Zhao Z, Fan H. Elevated TFAP4 regulates lncRNA TRERNA1 to promote cell migration and invasion in gastric cancer. Oncol Rep. 2018;40:923–931. doi: 10.3892/or.2018.6466. [DOI] [PubMed] [Google Scholar]
- 25.Yuan J, Zeng J, Shuai C, Liu Y. TWSG1 is a novel tumor suppressor in gastric cancer. DNA Cell Biol. 2018;37:574–583. doi: 10.1089/dna.2018.4188. [DOI] [PubMed] [Google Scholar]
- 26.Ji CD, Wang YX, Xiang DF, Liu Q, Zhou ZH, Qian F, Yang L, Ren Y, Cui W, Xu SL, et al. Kir2.1 interaction with Stk38 promotes invasion and metastasis of human gastric cancer by enhancing MEKK2-MEK1/2-ERK1/2 signaling. Cancer Res. 2018;78:3041–3053. doi: 10.1158/0008-5472.CAN-17-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.George JT, Jolly MK, Xu J, Somarelli J, Levine H. Survival outcomes in cancer patients predicted by a partial EMT gene expression scoring metric. Cancer Res. 2017;77:6415–6428. doi: 10.1158/0008-5472.CAN-16-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Sayed IY, Daher A, Destouches D, Firlej V, Kostallari E, Maillé P, Huet E, Haidar-Ahmad N, Jenster G, de la Taille A, et al. Extracellular vesicles released by mesenchymal-like prostate carcinoma cells modulate EMT state of recipient epithelial-like carcinoma cells through regulation of AR signaling. Cancer Lett. 2017;410:100–111. doi: 10.1016/j.canlet.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Skrypek N, Goossens S, De Smedt E, Vandamme N, Berx G. Epithelial-to-mesenchymal transition: Epigenetic reprogramming driving cellular plasticity. Trends Genet. 2017;33:943–959. doi: 10.1016/j.tig.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Dominguez C, David JM, Palena C. Epithelial-mesenchymal transition and inflammation at the site of the primary tumor. Semin Cancer Biol. 2017;47:177–184. doi: 10.1016/j.semcancer.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Silva SD, Alaoui-Jamali MA, Soares FA, Carraro DM, Brentani HP, Hier M, Rogatto SR, Kowalski LP. TWIST1 is a molecular marker for a poor prognosis in oral cancer and represents a potential therapeutic target. Cancer. 2014;120:352–362. doi: 10.1002/cncr.28404. [DOI] [PubMed] [Google Scholar]
- 32.Sun Y, Xu K, He M, Fan G, Lu H. Overexpression of Glypican 5 (GPC5) inhibits prostate cancer cell proliferation and invasion via suppressing Sp1-mediated EMT and activation of Wnt/β-catenin signaling. Oncol Res. 2017 Sep 6; doi: 10.3727/096504017X15044461944385. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi H, Du Y, Nakai K, Ding M, Chang SS, Hsu JL, Yao J, Wei Y, Nie L, Jiao S, et al. EZH2 contributes to the response to PARP inhibitors through its PARP-mediated poly-ADP ribosylation in breast cancer. Oncogene. 2018;37:208–217. doi: 10.1038/onc.2017.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nestal de Moraes G, Ji Z, Fan LY, Yao S, Zona S, Sharrocks AD, Lam EW. SUMOylation modulates FOXK2-mediated paclitaxel sensitivity in breast cancer cells. Oncogenesis. 2018;7:29. doi: 10.1038/s41389-018-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study available from the corresponding author on reasonable request.