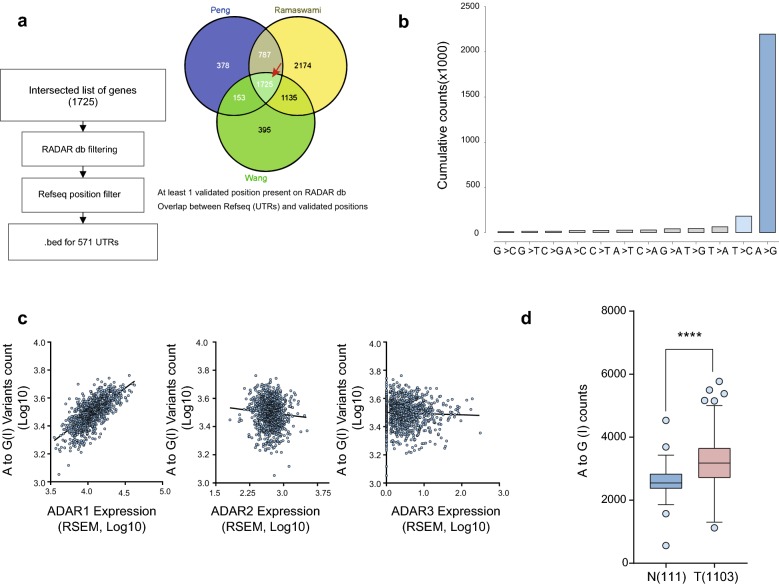
Fig. 2.
UTRs from the BRCA TCGA cohort show increased editing variants associated to ADAR1 activity. a Workflow for ADAR1-target selection, based on the intersection of three independent studies with complementary RNA editing study approaches. b Cumulative parameter count across the different possible variants across the UTRs analyzed. c Correlation of ADAR expression isoforms (RSEM, Log10) and the number of A to G(I) variants for each tumor (Log10). d A to G(I) counts in the 571 evaluated UTRs from BRCA TCGA cohort in normal and tumor breast samples. Box and Whiskers plot with Tukey distribution, ****: Kolmogorov–Smirnov p < 0.001. Pearson Correlation analysis for figure (c) r = 0.679 p < 0.0001 (ADAR1); r = − 0.077 p < 0.01 (ADAR2); r = − 0.028 p < 0.3765 (ADAR3)