Table 1. SFX data-collection and refinement statistics for data collected using the SOS sandwich.
Values in parentheses are for the highest resolution shell.
| Lysozyme | Hb.CO | |
|---|---|---|
| Data-collection statistics | ||
| Space group | P43212 | P212121 |
| a, b, c (Å) | 79.8, 79.8, 38.7 | 55.8, 157.4, 64.2 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| No. of crystals | 13802 [73% indexing rate] | 26039 [38% indexing rate] |
| Resolution range (Å) | 40–2.1 (2.20–2.10) | 39.3–2.2 (2.28–2.20) |
| Completeness (%) | 100 (100) | 99.96 (100) |
| Multiplicity | 283 (42) | 319 (65) |
| 〈I/σ(I)〉 | 10.3 (2.8) | 7.3 (2.2) |
| R split † | 0.090 (0.324) | 0.115 (0.466) |
| CC | 0.986 (0.773) | 0.980 (0.654) |
| CC* | 0.996 (0.934) | 0.995 (0.889) |
| Wilson B factor (Å2) | 33.3 | 42.3 |
| Refinement statistics | ||
| R/R free | 0.199/0.235 | 0.182/0.212 |
| R.m.s.d. | ||
| Bond lengths (Å) | 0.006 | 0.009 |
| Bond angles (°) | 0.962 | 0.980 |
| No. of atoms | ||
| Protein | 992 | 4332 |
| Water | 44 | 20 |
| Average B factors (Å2) | ||
| Protein | 31.2 | 43.5 |
| Water | 36.3 | 36.6 |
| Percentage of residues in areas of Ramachandran plot | ||
| Preferred | 96.0 | 98.9 |
| Allowed | 4.0 | 1.1 |
| Disallowed | 0.0 | 0.0 |
| PDB code | 6gf0 | 6hal |
R
split = 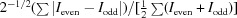 (White et al., 2012 ▸), where I
even and I
odd are intensities determined from all even-numbered and all odd-numbered images, respectively.
(White et al., 2012 ▸), where I
even and I
odd are intensities determined from all even-numbered and all odd-numbered images, respectively.
