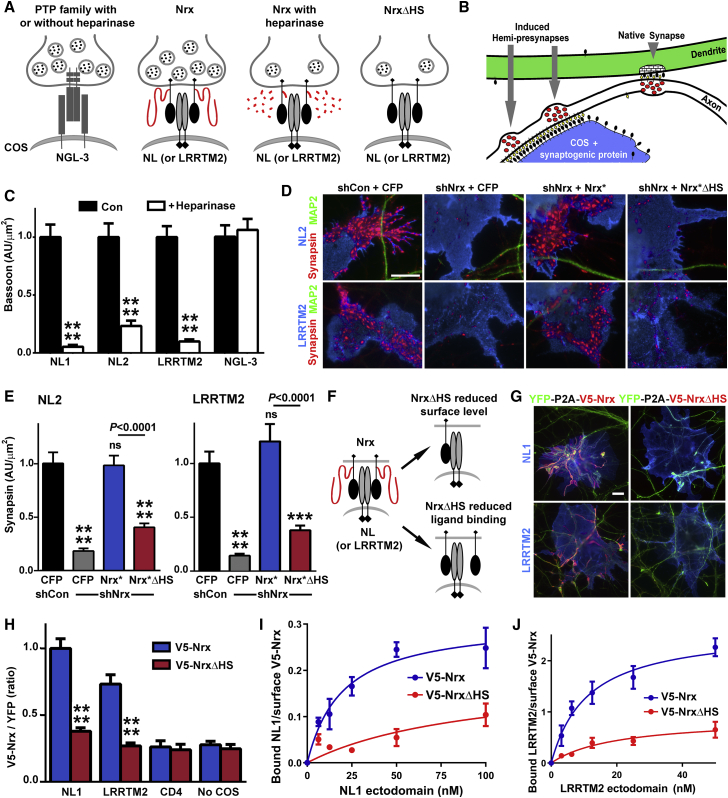
Figure 3.
Nrx HS Modification Is Required for Presynaptic Differentiation by NLs and LRRTM2
(A and B) Schematic models for panels (C)–(E).
(C) Clustering of presynaptic marker bassoon in contacting axons induced by Nrx ligands NL1, NL2, or LRRTM2 on COS7 cells, but not by NGL-3 a ligand of type IIA protein tyrosine phosphatases, was reduced by heparinase. Heparinase was added after axon outgrowth only during the co-culture period. Measures are integrated intensity of bassoon puncta, lacking dendrite contact to exclude native synapses, per transfected COS7 cell contact area.
(D and E) Clustering of presynaptic marker synapsin induced by NL2 or LRRTM2 was abolished by Nrx knockdown (shNrx) and rescued by RNAi-resistant Nrx∗ but not Nrx∗ΔHS. Measures are integrated intensity of synapsin puncta per transfected COS7 cell-axon contact area lacking MAP2 dendrite contact.
(F) Potential mechanisms by which Nrx HS modification might control synapse development.
(G and H) HS modification of Nrx regulates its recruitment by NL1 and LRRTM2. Neurons were transfected for YFP-P2A-V5-Nrx or YFP-P2A-V5-NrxΔHS and co-cultured with COS7 cells expressing NL1, LRRTM2, or control CD4. Measures are integrated intensity of V5-Nrx or V5-NrxΔHS per contact area of YFP-positive axons with transfected COS7 cells lacking dendrite contact (no COS7 indicates intensity on YFP-positive axons not contacting COS7 cells).
(I and J) HS modification of Nrx regulates interaction with NL1 and LRRTM2. Binding of NL1 or LRRTM2 ectodomain was measured on immature neurons expressing V5-Nrx or V5-NrxΔHS. For NL1 (I), scatchard analysis of this cell-based binding revealed a significant difference (p < 0.0001) with apparent Kd 19.7 and Bmax 0.31 for Nrx and Kd 95.2 and Bmax 0.19 for NrxΔHS. For LRRTM2 (J), scatchard analysis revealed a significant difference (p < 0.0001) with apparent Kd 11.1 and Bmax 2.6 for Nrx and Kd 19.8 and Bmax 0.9 for NrxΔHS.
∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001 by Kruskal-Wallis and Dunn’s tests comparing each heparinase with Con (C, n = 28–33) or compared to shCon+CFP (E, n = 39–55) or comparing each V5-NrxΔHS to V5-Nrx (H, n = 40–57). Error bars represent SEM. Scale bars: 10 μm. See also Figure S4.