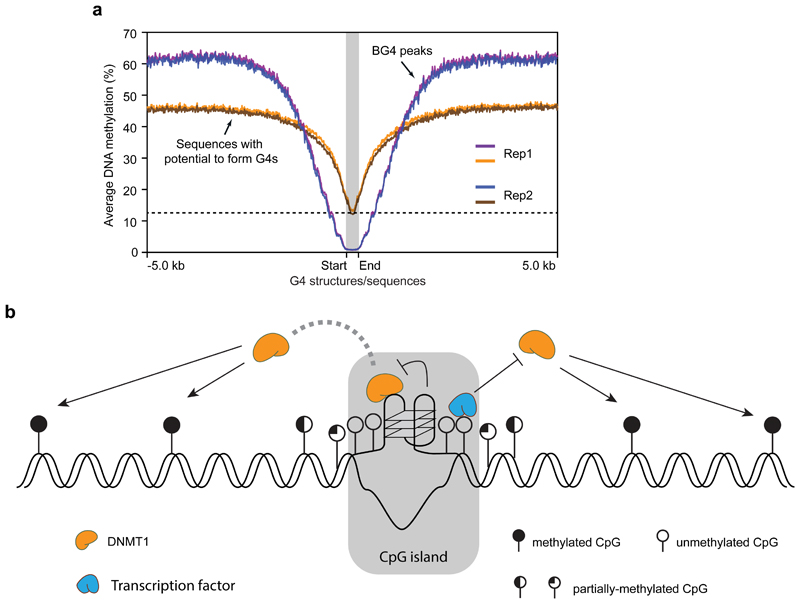
Figure 4. Recruitment of DNMT1 by G4 structures shapes the methylome in G-rich regions.
a) Plot showing the average methylation profile centred around G4 forming regions (red and blue are replicates 1 and 2 respectively, n = 7,491) or G4 sequences without structure (orange and green are replicates 1 and 2 respectively, n = 36,015). The plot extends ± 5 Kb from the centre. The dotted line denotes the lowest methylation level of G4 sequence without structure. b) Proposed model for potential involvement of G4 structures and methylation control at CGIs: i) G4 structures sequester DNMT1 due to high affinity binding; ii) G4 structures inhibit the methylation activity of DNMT1. Together with the binding of transcription factors, G4 structures contribute to protection of CGIs from methylation.