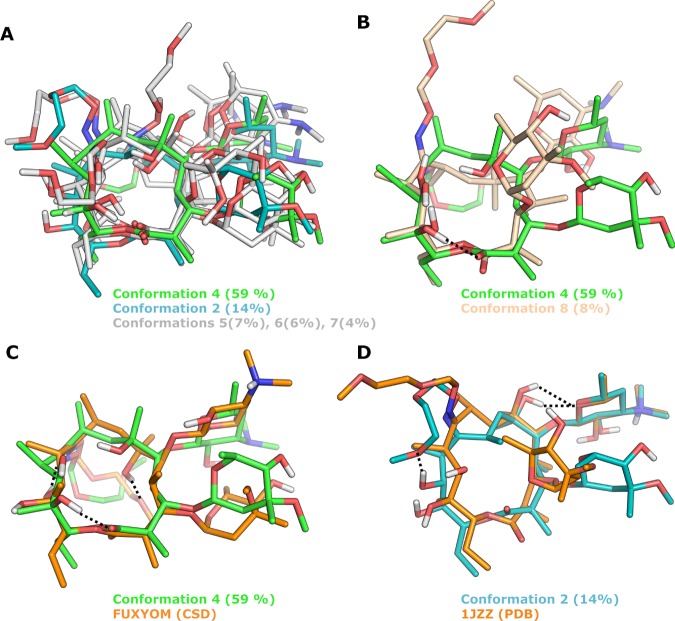
Figure 9.
Solution ensemble of roxithromycin in D2O, as determined by NAMFIS analysis and comparisons to two of the crystal structures of roxithromycin. (A, B) Overlays of the six conformations found in D2O, with the most populated one in green (number 4). For clarity, conformation 4 has been compared to four of the conformations in (A) and to one of the most different ones in (B). (C) Overlay of the most populated conformation (number 4, green) and the most similar crystal structure (CSD FUXYOM, orange); RMSD = 2.93 Å. (D) Overlay of solution conformation 2, with the crystal structure (PDB 1JZZ, orange) that is most similar to any of the six solution conformations; RMSD = 2.02 Å. Hydrogen bonds to the oxime side chain of roxithromycin are indicated by black dotted lines in (C) and (D), and nonpolar hydrogen atoms have been omitted for clarity in (A)–(D).