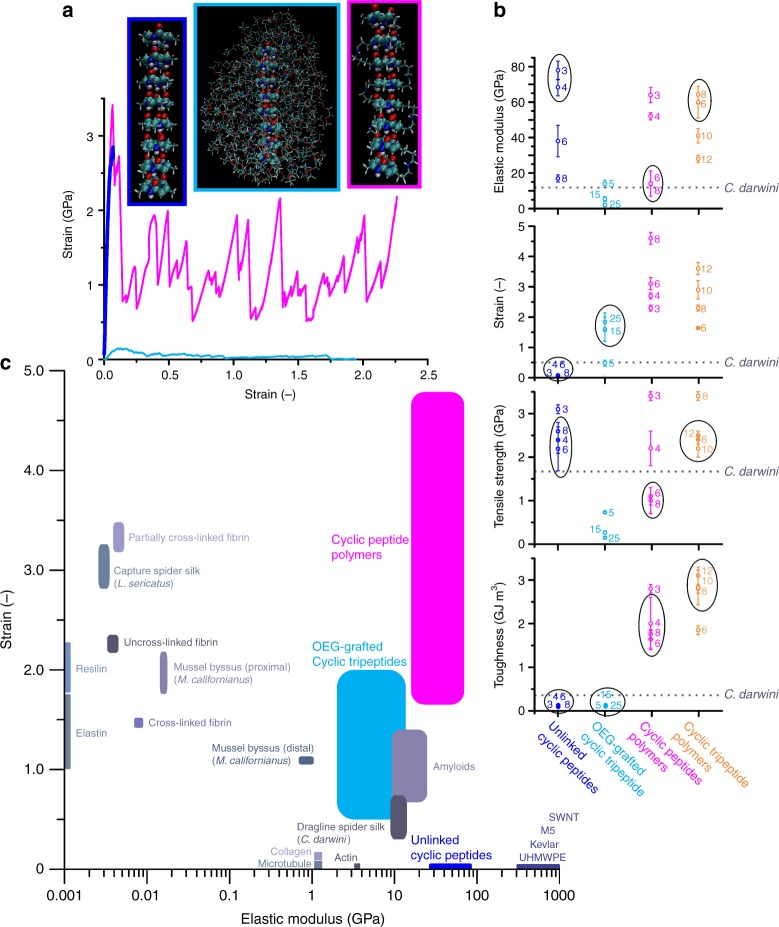
Fig. 1.
Molecular dynamics simulations of cyclic peptide assemblies. a Representative stress-strain curves of unlinked (cyclo[(-β-Ala)3-]; blue), OEG-grafted ((CH2CH2O)25CH3; cyan), and polymerized (cyclo[(-β-HLys-β-Ala-β-HLys)-] + cyclo[(-β-HGlu-β-Ala-β-HGlu)-]; magenta) assemblies consisting of eight subunits. In MD simulations, α-carbon atoms in the backbone of the bottom rings are held fixed, while a spring with a spring constant of 5000 kJ mol−1 is attached to the center of mass of the top ring and pulled upward along the fibril axis at a constant velocity of 5 × 10−5 nm ps−1. b Computed elastic moduli, strain to failures, tensile strengths, and toughness values for the various cyclic peptide based assemblies (mean ± standard deviation (SD); N = 3). Dotted lines denote the reported values of dragline spider silk from C. darwini8. Numbers next to data points indicate the number of amino acids in each subunit (blue and magenta), number of OEG repeating units (cyan), or number of atoms in the linkage between subunits (orange). Circles denote data pairings that are not statistically different based on unpaired t tests (p < 0.05). c Computed elastic moduli and strain to failures of cyclic β-peptide assemblies, in comparison with reported values for natural and synthetic materials4, 8, 9, 17, 34, 54–60. Single-walled carbon nanotubes are abbreviated as SWNT, and ultra-high molecular weight polyethylene is abbreviated as UHMWPE