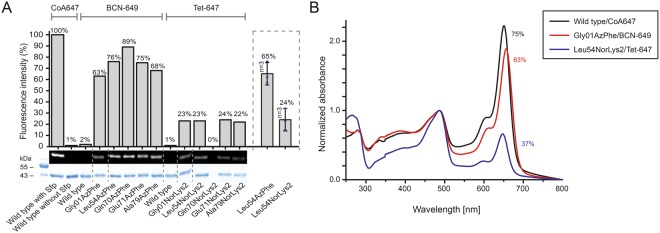
Figure 5.
Fluorescent labelling of ACP-GFP mutants. (A) DOL of ACP-GFP mutants in respect to the amber mutation site determined by relative in-gel fluorescence intensities at wavelength 650 nm. The ACP-GFP construct was enzymatically modified by a fluorescent CoA647-label with Sfp and served as the wild type reference. Hence, it was put to 100% fluorescence intensity. AzPhe mutants were labelled with 80 equiv. of BCN-POE3-NH-DY649P1 (BCN-649), NorLys2 mutants were labelled with 80 equiv. of 6-methyl-tetrazine-ATTO-647N (Tet-647) in 10 µL reaction volume. All fluorescence intensities were corrected by the quantum efficiency of the respective fluorophore and correlated to the protein bands of the Coomassie-stained gel (lanes have been assembled for clarity, as indicated by dashed lines). Scans of the original gels are presented in Supplementary Fig. S10. Biological replicates were performed for the Leu54 mutants, comparing DOL determined for three parallel labelling reactions analysed on different fluorescent gels (inserted box). Individual results were gathered from gels in A and Supplementary Fig. S4. (B) DOL determined by UV-Vis spectroscopy. 25 equiv. of fluorophore were used in labelling reactions of ACP-GFP mutants in 50 µL reaction volume. Free fluorophore was removed by purification over Ni-NTA magnetic beads. UV-Vis absorbance spectra were normalized to GFP absorbance at wavelength 485 nm. DOL is read out by comparing absorbance of the fluorophore at 650 nm to absorbance of GFP at 485 nm.