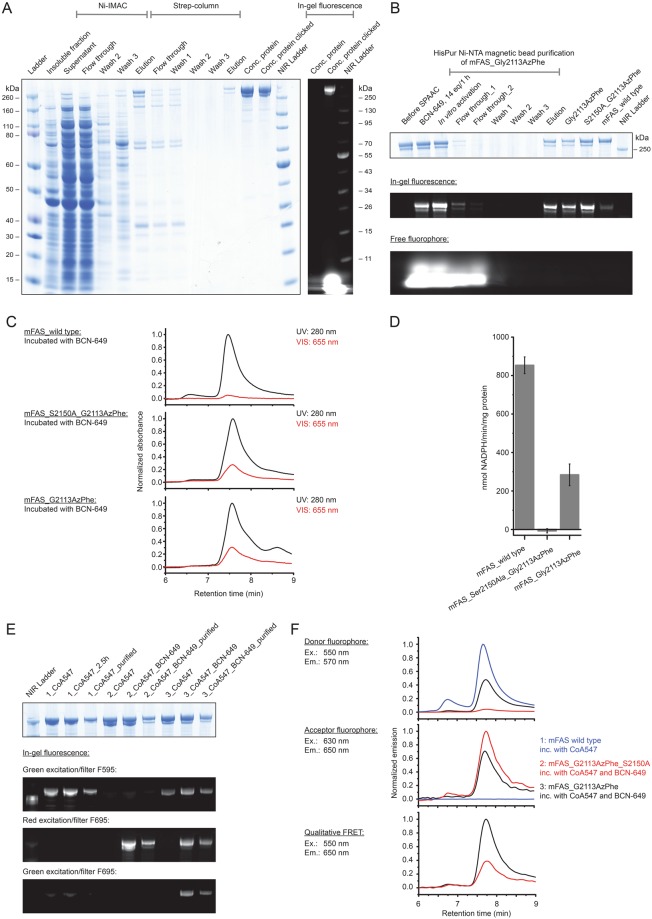
Figure 6.
Physicochemical analysis of fluorescently labelled mFAS. (A) SDS-PAGE (NuPAGE Bis-Tris 4–12%) of a representative purification of the ncAA-modified mFAS mutant (Gly2113AzPhe with additional ACP knock-out mutation Ser2150Ala). The portion of truncated polypeptide chains after the tandem purification strategy was quantified to roughly 30%. Truncated proteins reflect insufficient amber suppression and are co-purified by heterodimer formation. In-gel fluorescence demonstrates successful labelling with fluorophore BCN-649. (B) Purification of three full-length mFAS variants (Gly2113AzPhe, Gly2113AzPhe with Ser2150Ala, and wild type) in preparative 100 µg scale via HiPur Ni-NTA magnetic beads. Samples were clicked with 16 equiv. BCN-649 for 1 h followed by in vitro phosphopantetheinylation with CoA and Sfp. In-gel fluorescence of a SDS-PAGE (Bis-Tris 4–12%) was detected before Coomassie-staining. The lowest panel indicates that the majority of free fluorophore is washed away after the first washing step. (C) Analysis of the integrity of the three labelled samples after clicking and purification by HPLC-SEC. The main peak at 7.5–7.6 min corresponds to the native dimeric state. Absorbance was monitored at 280 nm and 655 nm and normalized to the highest peak in the UV signal. Both samples containing the ncAA AzPhe were labeled with fluorophore (28% and 31%), whereas the wild type mFAS showed only minor non specific fluorophore binding (5%). (D) Activity of mFAS variants monitored by a NADPH consumption assay after phosphopantetheinylation and clicking. The variant Gly2113AzPhe showed one third of the wild type activity, whereas the ACP knock-out (Ser2150Ala) could not produce fatty acids. Error bars indicate the technical repeatability determined from three repeated experiments per construct. (E) SDS-PAGE (NuPAGE Bis-Tris 4–12%) of the same three variants: 1: wild type mFAS, 2: mFAS (Gly2113AzPhe and Ser2150Ala) and 3: mFAS (Gly2113AzPhe) after enzymatic labelling with CoA-547 and clicking of the AzPhe containing variants with BCN-649. In-gel fluorescence was detected with three different settings: excitation with green light and filter F595 (for 595 nm); excitation with red light and filter F695 (695 nm) and excitation with green light and filter F695. The gel after Coomassie-staining is attached. All samples show specific fluorescent bands in the respective channels with little unspecific binding due to denaturing conditions. The doubly labelled sample Gly2113AzPhe shows FRET signal. (F) Fluorescence analysis of the three labelled samples of (E) via HPLC-SEC. Emission spectra are shown for the settings: Ex. 550 nm/Em 570 nm; Ex. 630 nm/Em 650 nm and Ex. 550 nm/Em 650 nm. All signals were normalized as described in detail in the methods section. Again, the doubly labelled sample Gly2113AzPhe shows (highest) FRET signal.