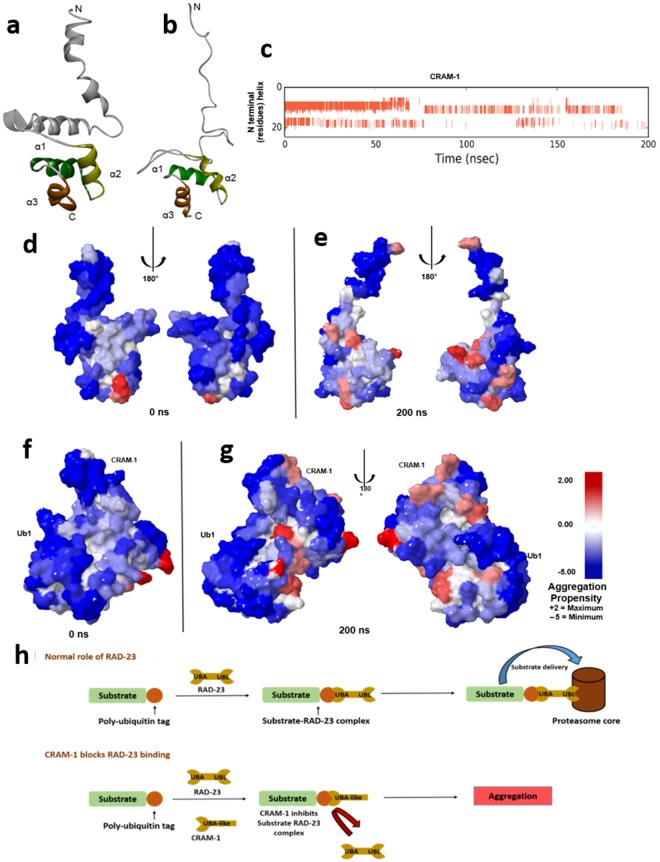
Figure 5.
Atomistic molecular-dynamic simulation predicts aggregation-prone regions in CRAM-1 due to structural unfolding. (a,b) MD simulation in Desmond (Desmond Molecular Dynamics System, ver. 2016.4, D.E. Shaw Research, New York, NY) showing the unfolding of protein structure from initial (a) to simulation-stability (b) conformations. (c) Helical regions (red bars) indicate unfolding of N-terminal residues, plotted against simulation time (x axis). (d–g) Predicted aggregation propensity (see scale at right) of CRAM-1 alone before simulation of structural rearrangement (d), or at the end of simulation (e) and of CRAM-1 bound to mono-ubiquitin (ub1) before simulation (f), or at the end of simulation (g). (h) Schematic depiction of a proposed mechanism, wherein CRAM-1 competes with RAD-23 for binding to ubiquitin, thus impeding clearance of ubiquitinated substrates.