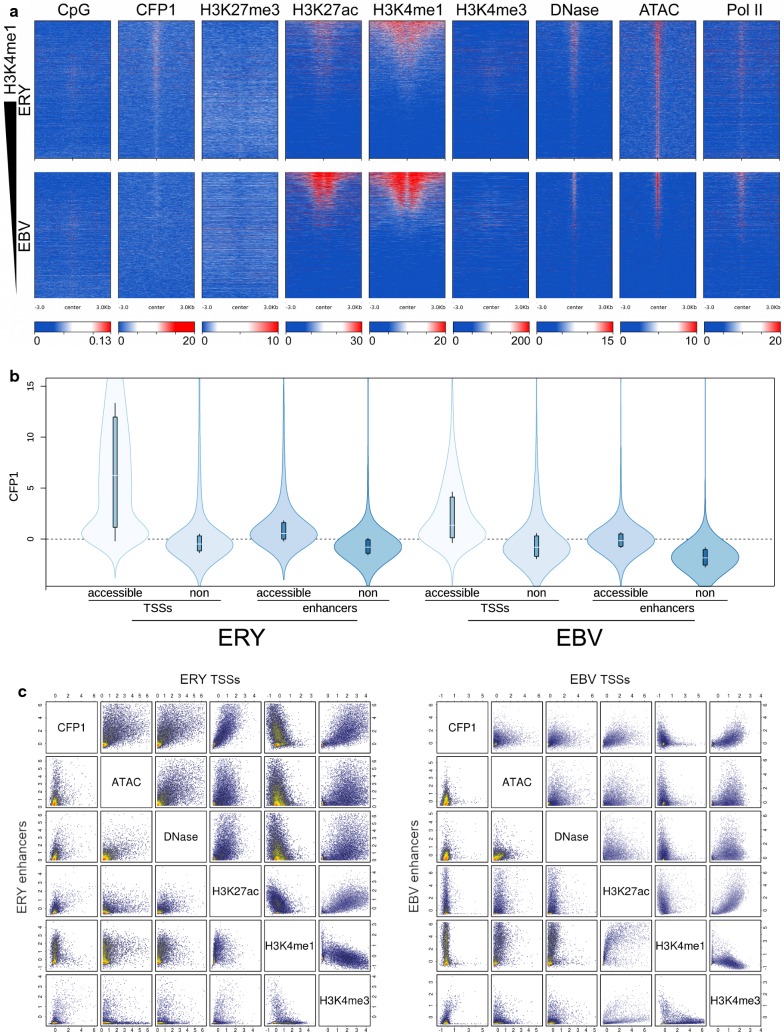
Fig. 4.
CFP1 binding and histone methylation marks in putative enhancers and comparison to TSSs. a Loci sorted by H3K4me1 signal in 6-kb regions surrounding putative enhancers. Upper panels, ERY; lower, EBV. Note that all data sets, other than CFP1, differ by source between ERY and EBV, which explains prominent differences in signal intensities, for example, of H3K27ac and H3K4me1. b Quantitative comparison of signal intensity in TSSs and putative enhancers, above and below median accessibility (as determined by ATAC-seq data), with cell types indicated. c Plots of ChIP signals for erythroid (left) and lymphoid cells (right), in putative enhancers (lower left graphs) and TSSs (upper right graphs)