Figure 2.
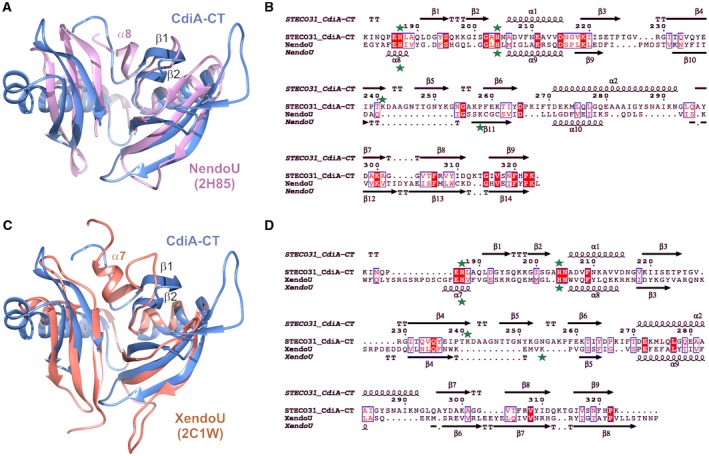
The CdiA‐CTSTECO31 toxin is a prokaryotic EndoU domain. A. CdiA‐CTSTECO31 (blue) and the C‐terminal domain of SARS Nsp15 (PDB: 2H85, pink) were aligned using secondary structure matching (SSM) superposition calculated in Coot (Emsley and Cowtan, 2004). The β1‐β2 hairpin of CdiA‐CTSTECO31 and helix α8 in the NendoU active‐site loop are indicated. B. The sequences of the CdiA‐CTSTECO31 and SARS Nsp15 C‐terminal domains were aligned based on structure using DALI (Holm and Rosenstrom, 2010). The resulting sequence alignment was rendered using Espript (Robert and Gouet, 2014). Identical residues are highlighted in red, and similar residues are shown in red font. The predicted catalytic triad residues are marked with green stars. C. CdiA‐CTSTECO31 (blue) and the C‐terminal domain of XendoU (PDB: 2C1W, coral) were aligned by SSM superposition. The β1‐β2 hairpin of CdiA‐CTSTECO31 and helix α7 in the XendoU active‐site loop are indicated. D. CdiA‐CTSTECO31 and XendoU sequences were aligned as described in panel B. [Colour figure can be viewed at http://wileyonlinelibrary.com]
