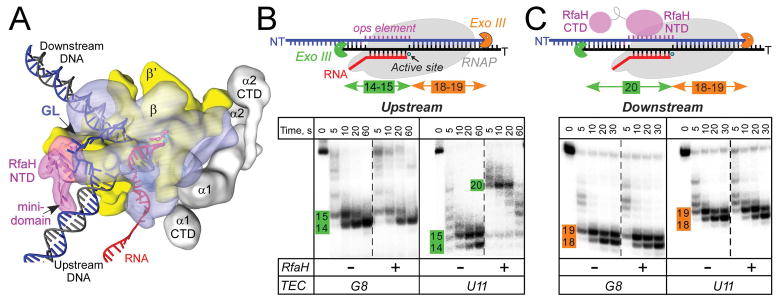
Figure 1.
RfaH interactions with the EC.
A. A model of the RfaH-bound EC. RNAP α (grey), β (blue), and β′ (yellow) subunits and RfaH NTD (magenta) are depicted by simplified molecular surfaces; β (with the GL labeled) and RfaH NTD are rendered semi-transparent. Nucleic acids are shown as cartoons, two Mg2+ ions in the active site – as cyan spheres, and an incoming NTP – as red sticks.
B and C. Footprinting of the upstream (B) and downstream (C) RNAP boundary in G8 and U11 ops ECs. Exo III was added to ECs assembled on synthetic scaffolds in which RNAP is halted at G8 and U11 positions in the ops element (NT sequence G1GCGGTAG8CGT11G); the probed DNA strand (T, panel B; NT, panel C) was 5′-end labeled with [γ32P]-ATP. RfaH was present at 100 nM where indicated. Aliquots were quenched at the indicated times (0 represents an untreated DNA control) and analyzed on a 12 % denaturing gel; a representative of three independent experiments is shown. Numbers indicate the distance from the RNAP active site.