Figure 3. Altered translation of alternative splicing factors and activation of a neonatal splicing program in regenerating hepatocytes.
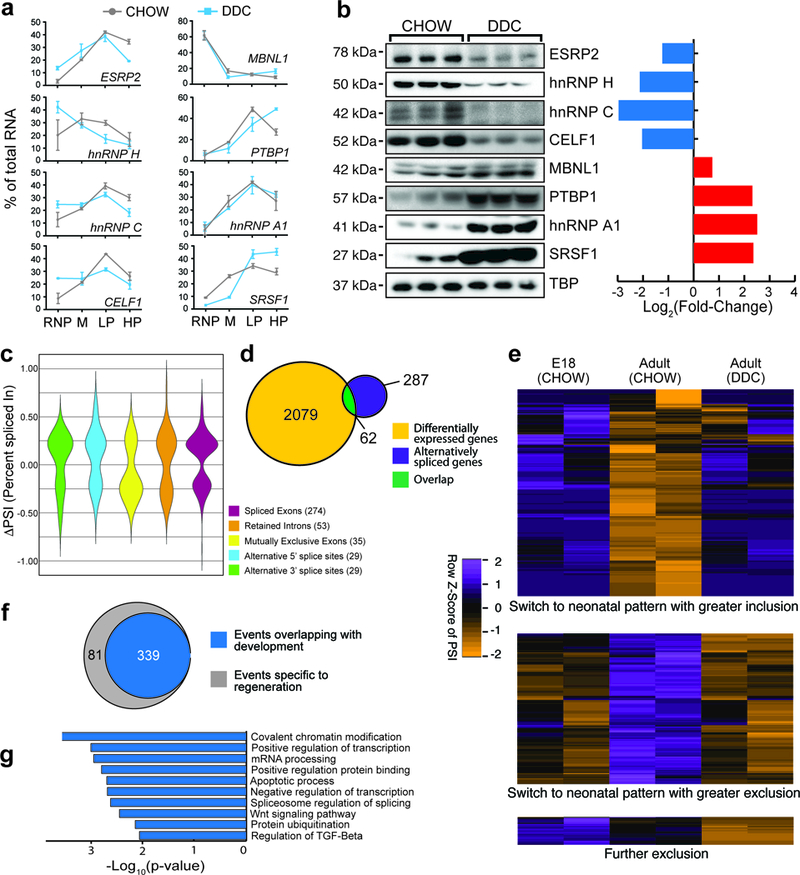
a,qPCR-based expression of select alternative splicing factors in different polysomal fractions of hepatocytes isolated from CHOW and DDC-fed mouse livers (n = 3 animals/condition). M: monosome, LP: Light polysome, HP: Heavy polysome. Data are mean ± s.d. b, Western blots and their respective quantification showing changes in protein abundance of select alternative splicing factors (n=5 animals, all blots were repeated independently at least 3 times). c, Violin distribution plots for significantly changing alternative splicing events after DDC injury according to the event types (p<0.05, FDR<0.10 from rMATS: adjusted for multiple testing, Difference in Percent Spliced Index [ΔPSI]≥15%, and Junction Counts≥10) obtained from RNA-seq of 2 biological replicates for each condition. d, Overlap of mRNA abundance and alternative splicing changes following DDC injury. e, Heatmap of PSI values showing splicing transitions (row normalized, PSI) overlapping between development (E18 CHOW-Adult CHOW) and regeneration (Adult CHOW-Adult DDC). Orange color indicates reduced exon inclusion and purple color indicates higher exon inclusion following DDC injury. Each paired column is an independent biological replicate. f, Venn diagram showing number of alternative splicing events in regenerating hepatocytes that are reciprocally regulated during normal development. g, GO analysis for significantly enriched pathways and molecular functions for genes undergoing alternative splicing following DDC injury. All uncropped gel images are provided in Supplementary data set 1.
