Figure 4. Regeneration-regulated alternative exons are enriched in unstructured protein domains and phosphorylation sites. a.
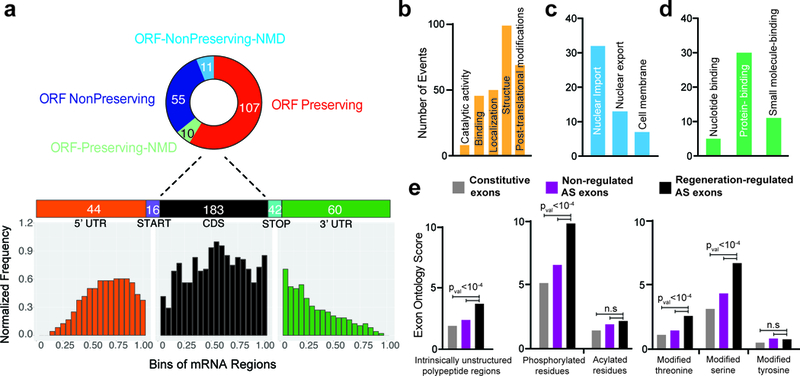
a,Breakup of DDC injury-induced alternatively spliced (AS) events into relative transcript regions, and their effects on the open reading frame (top). Metagene analysis of differentially spliced exons by their position on an mRNA transcript (bottom). b-d, Exon Ontology based distribution of differentially spliced exons according to the encoded protein features (b) and sub-distribution by localization (c), and binding (d). e, Regeneration-regulated alternative exons code for protein segments enriched for unstructured polypeptide regions and experimentally confirmed phosphorylated residues especially phospho-serine and phospho-threonine residues. Sample size is 247 regeneration-regulated mouse cassette exons. Background set and statistics (t-test, two sided) were used as defined by the ExonOntology and FasterDb databases. n.s: not significant.
