Figure 6. ESRP2 downregulation reprograms alternative splicing to generate neonatal isoforms of Hippo pathway proteins. a.
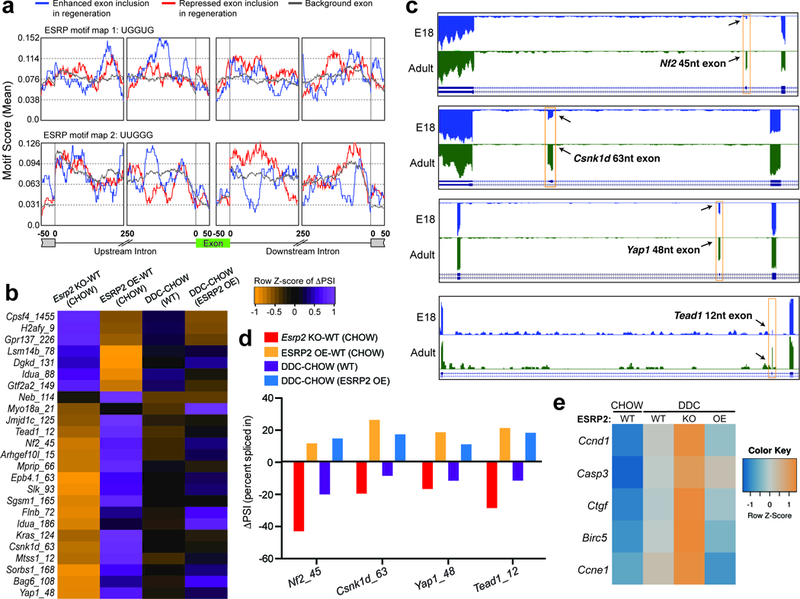
a,Position and relative enrichment of ESRP2 binding motifs within exonic and 250nt intronic sequences surrounding the regeneration-regulated exons. b, Heatmap of differences in RT-PCR derived exon inclusion (row-normalized, ΔPSI) values in WT, Esrp2KO and ESRP2 overexpression (OE) livers under CHOW and DDC conditions (n=3). Numeral after gene name signifies the size of alternative exon. c, Genome browser views of RNA-seq data from E18 and adult mouse hepatocytes demonstrating developmental regulation of alternative exons in Hippo pathway genes-Nf2, Csnk1d, Yap1 and Tead1 (n= 2 biological replicates). d, Difference in Percent Spliced Index (ΔPSI) values for Nf2, Csnk1d, Yap1 and Tead1 alternative exons (n=3), and e, Heatmap of relative expression for Hippo target genes (qPCR) in WT, Esrp2KO and ESRP2 OE livers under CHOW and DDC conditions (n=3).
