Abstract
The genetic causes of many Mendelian disorders remain undefined. Factors such as lack of large multiplex families, locus heterogeneity, and incomplete penetrance hamper these efforts for many disorders. Previous work suggests that gene-based burden testing—where the aggregate burden of rare, protein-altering variants in each gene is compared between case and control subjects—might overcome some of these limitations. The increasing availability of large-scale public sequencing databases such as Genome Aggregation Database (gnomAD) can enable burden testing using these databases as controls, obviating the need for additional control sequencing for each study. However, there exist various challenges with using public databases as controls, including lack of individual-level data, differences in ancestry, and differences in sequencing platforms and data processing. To illustrate the approach of using public data as controls, we analyzed whole-exome sequencing data from 393 individuals with idiopathic hypogonadotropic hypogonadism (IHH), a rare disorder with significant locus heterogeneity and incomplete penetrance against control subjects from gnomAD (n = 123,136). We leveraged presumably benign synonymous variants to calibrate our approach. Through iterative analyses, we systematically addressed and overcame various sources of artifact that can arise when using public control data. In particular, we introduce an approach for highly adaptable variant quality filtering that leads to well-calibrated results. Our approach “re-discovered” genes previously implicated in IHH (FGFR1, TACR3, GNRHR). Furthermore, we identified a significant burden in TYRO3, a gene implicated in hypogonadotropic hypogonadism in mice. Finally, we developed a user-friendly software package TRAPD (Test Rare vAriants with Public Data) for performing gene-based burden testing against public databases.
Keywords: hypogonadotropic hypogonadism, gene-based burden analysis, TRAPD
Introduction
In the past, most gene discovery for rare disorders was performed using linkage analysis in large families. However, the advent of next-generation sequencing (NGS) has enabled alternative statistical approaches to gene discovery including a gene-based burden testing approach. In this approach, the number of individuals carrying rare, protein-altering variants in each gene is compared between case and control subjects. The motivation behind this approach is that while a single variant is usually underpowered to detect statistical signals between case and control subjects, aggregating variants across a candidate gene might improve power. This approach has a further advantage that it can be applied to unrelated case subjects, thus overcoming some limitations of linkage analysis, including for disorders where there are a lack of large multiplex families or where there is considerable incomplete penetrance.1 This approach has already led to the discovery of a number of genes associated with various disorders and has been also referred to as gene-based collapsing analysis.2 It is highly similar to burden testing approaches applied in rare variant association studies (RVASs), where sums of allele counts of rare protein-altering variants in each gene are compared between case and control subjects.3, 4, 5
Recently, large exome-sequencing databases have been generated, including the Exome Aggregation Consortium (ExAC) and the Genome Aggregation Database (gnomAD).6 To date, these databases have been used largely as variant frequency databases to filter out more common variants that are unlikely to be pathogenic for a Mendelian disorder. These databases also have the potential to serve as controls in burden-testing strategies, but there are important limitations to the use of these databases for this purpose. First, only variant-level data are typically released, thus making it challenging to perform more sophisticated tests such as SKAT that require individual-level genotype data.3 Second, these databases aggregate data obtained from multiple sequencing platforms, which may differ from the platform(s) used to sequence case subjects. Third, public database samples are not jointly processed and variant-called with the case samples, potentially introducing additional technical artifacts. Finally, although public databases provide some ancestry information, the exact ancestry of the individuals is not available and hence cannot be readily and directly compared to the ancestry of the case subjects.
This study leverages gnomAD, the largest publicly available sequencing cohort (n = 123,136) as control subjects, to perform burden testing against 393 individuals with idiopathic hypogonadotropic hypogonadism (IHH) for whom whole-exome sequencing was performed. IHH is a rare Mendelian disorder characterized by the failure of normal pubertal development due to deficiency of gonadotropin-releasing hormone (GnRH) and has a prevalence of approximately 1:30,000–1:125,000.7, 8 To date, approximately 35 genes have been associated with IHH.9, 10 However, these genes account for only approximately one-third of cases of disease, leaving the genetic basis of the disease unknown in the majority of affected individuals.
Given the reproductive consequences of this disorder, a fully penetrant IHH-causing mutation cannot be passed down from an affected individual to their offspring. Thus, most individuals with IHH are simplex cases, or the IHH-causing mutations are incompletely penetrant. Both factors hamper linkage-based approaches for IHH. Previous work suggests that a gene-based burden approach, which is capable of leveraging unrelated probands, would have increased power to uncover new genes as compared to traditional family-based approaches.1
To overcome the dual obstacles of using public control data and of gene discovery in a rare Mendelian disease cohort, this study utilizes a variety of approaches to harmonize case and control data. Synonymous variants, which are presumably largely benign, are used to calibrate this approach to guard against type I error. The 35 genes previously implicated in IHH are used as positive controls to evaluate the efficacy of the approach. Approaches to overcome the various pitfalls of conducting rare variant gene-based burden testing using public control data are demonstrated through sequential evaluations of performance metrics.
These methods demonstrate that even in a rare disorder with substantial locus heterogeneity such as IHH, it is possible to rediscover known genes as well as validate candidate genes. This approach, available in a new user-friendly software package TRAPD (Test Rare vAriants with Public Data), can be used by other rare disease researchers for gene discovery and validation efforts.
Subjects and Methods
Recruitment
The study was approved by the Institutional Review Board at the Massachusetts General Hospital (MGH). All study individuals or their legal guardians provided written informed consent. Individuals with IHH have been referred to and/or recruited by the Reproductive Endocrine Unit (REU) at MGH over 20 years. IHH was defined as hypogonadal sex-steroid levels (testosterone < 100 ng/dL in men; estradiol < 20 pg/mL in women) in the setting of low or normal gonadotropin levels at age ≥18 years and the absence of any identifiable medical condition that could cause hypogonadotropic hypogonadism. Anosmia was demonstrated either by formal smell testing using the University of Pennsylvania Smell Identification Test or a self-reported inability to smell (which has been demonstrated to correlate well with formal smell testing).11
Sample Ascertainment and Sequencing
Whole-exome sequencing was performed from peripheral blood-derived DNA in three separate batches. The first batch (n = 100; 60 case subjects) was sequenced at the Yale Center for Mendelian Genomics (New Haven, CT), with exome capture performed using the Nimblegen SeqCap target enrichment kit (Roche). The second batch (n = 161; 143 case subjects) was sequenced at the Broad Institute (Cambridge, MA), with exome capture performed using the Agilent SureSelect v2 capture kit (Agilent Technologies). The third batch (n = 1,076; 190 case subjects) was sequenced at the Broad Institute with exome capture performed using a custom Illumina capture kit (ICE) (Illumina). These sequencing batches contained additional samples that were not a part of this study.
For the first batch (sequenced at Yale) and third batch (sequenced at Broad using ICE capture), consecutive individuals in the MGH REU collection were selected. For the second batch of individuals (sequenced at the Broad Institute using SureSelect v2 capture), samples were selected in two ways. First, individuals with IHH bearing known genetic causes of IHH were screened out based on Sanger sequencing-based screening for 14 genes implicated in IHH: CHD7 (MIM: 608892), FGF8 (MIM: 600483), FGFR1 (MIM: 136350), GNRH1 (MIM: 152760), GNRHR (MIM: 138850), HS6ST1 (MIM: 604846), ANOS1, previously called KAL1 (MIM: 300836), KISS1 (MIM: 603286), KISS1R (MIM: 604161), NSMF, previously called NELF (MIM: 60813), PROK2 (MIM: 607002), PROKR2 (MIM: 607123), TAC3 (MIM: 162330), and TACR3 (MIM: 162332). Second, the batch was enriched for individuals with IHH bearing heterozygous mutations in PROKR2, which is classically implicated in IHH in the bi-allelic state.12 All case subjects were ascertained from different families, and this was confirmed by ensuring that identity by descent was <0.05 between all individuals.
Variant Calling and Annotation
Following sequencing, the resulting reads were aligned to the hg19 reference genome with BWA,13 applied GATK v.3.2,14 base quality score recalibration, indel realignment, and duplicate removal, and we performed SNP and indel discovery and genotyping across all samples simultaneously using standard hard filtering or variant quality score recalibration according to GATK Best Practices recommendations.15, 16 For purposes of burden testing, statistical genotypes were converted to discrete alternate allele counts (0, 1, or 2 representing homozygous reference, heterozygous, or homozygous variant, respectively). Variant call files (VCF) files were processed using Tabix v.1.317 and Bcftools v.1.2.18
Following variant calling, variants were annotated for functional effect using Variant Effect Predictor v.77.19 Three separate protein-prediction algorithms, PolyPhen2,20 SIFT,21 and CADD,22 were employed. Variants were also annotated for allele frequencies from TOPMed Freeze 5 and gnomAD v.2.0.2 (see Web Resources). For gnomAD, minor allele frequency (MAF) from each of the ancestries within gnomAD (African, Admixed American, Ashkenazi Jewish, East Asian, Finnish, non-Finnish European, and South Asian) were added and MAF filtering was based on the highest MAF from each of the gnomAD populations (i.e., population maximum allele frequency).
Adjusting for Read Depth
Each coding exon was annotated per GENCODE v.19. A 10 bp pad was included on either end of each exon. For each coding base, the proportion of individuals covered at >10× was calculated separately for IHH and gnomAD. Depth of coverage for each position for gnomAD samples was provided by the gnomAD consortium.6 Bases with at least 90% of samples covered in both IHH and gnomAD were retained for analyses. Finally, low-complexity repeats23 and segmental duplications (see Web Resources) were filtered out.
To apply the approach for adjusting for read depth used in Raghavan et al.24 (“Binomial Method”), at each coding base, the proportion of individuals covered at >10× in case cohort sequencing and gnomAD was calculated. The difference in these proportions for each site was then compared using a binomial test, and only sites that were not significantly different (p > 0.001) were retained for analyses. To apply the approach for adjusting for read depth used in Cirulli et al.2 (“Concordance Method”), at each coding base, the concordance of bases covered at >10× in greater than 90% of samples in case cohort sequencing as compared to gnomAD was calculated. Only exons with >90% concordance in bases meeting these criteria in case cohort sequencing versus gnomAD were retained.
Assessment of Quality by Depth (QD) and Variant Quality Score Log-Odds (VQSLOD) Filters
QD and VQSLOD filters were determined separately for case and control sequencing using a set of rare variants (MAF < 1.0%) that were shared between the case cohort and gnomAD. QD and VQSLOD filters were calculated at the 70, 75, 80, 85, 90, and 95th percentiles separately for the case cohort and gnomAD. Every pairwise combination of QD and VQSLOD filter between the case cohort and gnomAD was tested to establish the best match of synonymous variant burden testing between case and control datasets. This was done in similar fashion when comparing the case cohort to ExAC. Calibration of quality filters for indels was done similarly, except all rare nonsynonymous variants were used rather than synonymous variants.
PCA Analysis
Approximately 15,000 autosomal exonic SNPs from HapMap Phase 325 were used to conduct PCA analysis. SNPs were pruned using PLINK26 based on LD (variance inflation factor threshold of 2), and only common variants (MAF > 5%) were used in the PCA analysis. PCA outlier analyses were performed by projecting the case samples onto HapMap3 samples using EIGENSOFT.25, 27
Gene-Based Burden Testing
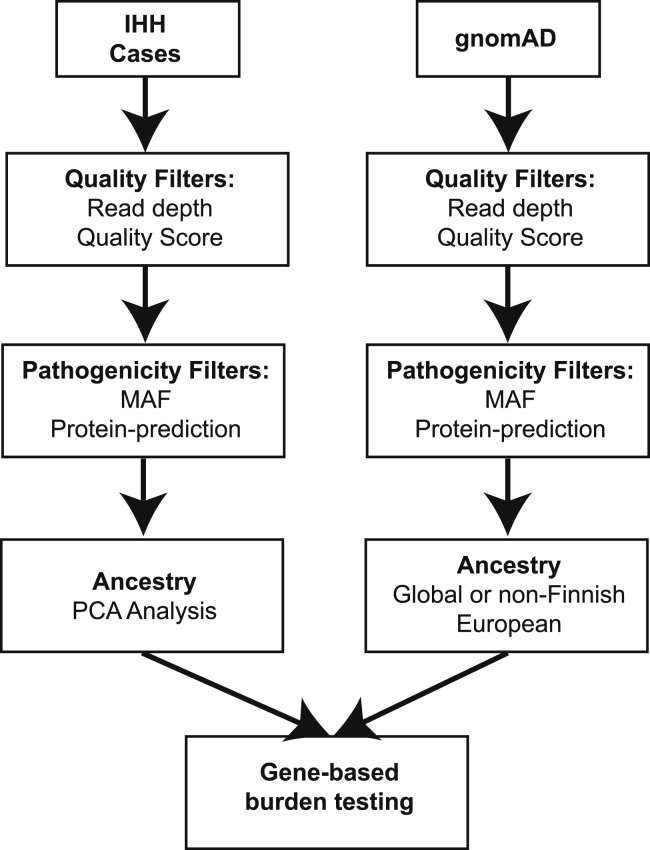
To perform gene-based burden testing, high-confidence variants and variants that are likely pathogenic based on MAF and/or protein-prediction algorithms were selected using various filters (specified in each individual section below). Variants that met these quality and pathogenicity filters are referred to as “qualifying variants” (Figure 1).2 A file listing the qualifying variants for each gene was generated using the “make_snp_file.py” function in TRAPD.
Figure 1.
Burden Testing Scheme
Case cohort sequencing (IHH) and control database sequencing (gnomAD) data are processed separately, and burden testing is performed in the final step. For each set of data, sequencing quality filters, predicted variant pathogenicity filters, and sample filters (e.g., ancestry) can be applied. Then, counts of qualifying variant carriers for each gene in the case and control subjects are generated. Finally, burden testing is performed.
For the dominant test, for each gene, the number of individuals in the case cohort who carry at least one qualifying variant in that gene was tabulated using the “count_cases.py” function in TRAPD. For the gnomAD control dataset, only summary statistics were available. Therefore, to approximate the number of control subjects carrying at least one qualifying variant in a given gene, the allele counts for all qualifying variants in that gene were summed. This approximation may be an overestimate if some individuals carry multiple variants in the same gene, an issue that is particularly salient in larger genes and/or those with a high rate of background variation. In this situation, the sum control allele counts would be inflated, resulting in a more conservative test. The tabulation of these counts for each gene was generated using the “count_controls.py” function in TRAPD.
For the recessive test, the case cohort counts were generated by tabulating the number of case subjects carrying two or more qualifying variants in each gene. Phase for variants in the case subjects cannot readily be determined; thus, some individuals carrying two variants in the same gene actually represent two variants on the same haplotype (and thus not bi-allelic). The control counts were generated by adding the number of individuals who harbor homozygous variants for each qualifying variant in a given gene to an estimated number of individuals who are compound heterozygotes. To estimate the number of individuals who are compound heterozygotes, the cumulative frequency of heterozygous variant carriers in a gene was squared and multiplied by the total number of individuals in gnomAD. This again may be an overestimate, which would result in a more conservative test.
Following tabulation of case and control counts, a 2 × 2 contingency table is generated for each gene. This contingency table represents the number of case and control subjects who carry and do not carry a qualifying variant in each gene. p values were calculated using two-sided Fisher’s exact test and p values < 2.5 × 10−6 were considered significant (α corrected for testing approximately 20,000 genes). Burden testing was performed using the “burden_test.R” function in TRAPD. Results were then visually represented by a quantile-quantile (QQ) plot.
Calibration with Genes Previously Implicated in IHH: “Positive Controls”
To serve as positive controls, a set of 35 autosomal genes previously implicated in IHH was analyzed: AXL (MIM: 109135), CHD7, DMXL2 (MIM: 612186), DUSP6 (MIM: 602748), FEZF1 (MIM: 613301), FGF8, FGFR1, FGF17 (MIM: 603725), FLRT3 (MIM: 604808), GLCE (MIM: 612134), GNRH1, GNRHR, HS6ST1, IL17RD (MIM: 606807), KISS1, KISS1R, KL (MIM: 604824), KLB (MIM: 611135), LEPR (MIM: 601007), NSMF, OTUD4 (MIM: 611744), PCSK1 (MIM: 162150), POLR3A (MIM: 614258), POLR3B (MIM: 614366), PROK2, PROKR2, RNF216 (MIM: 609948), SEMA3A (MIM: 603961), SOX10 (MIM: 602229), SPRY4 (MIM: 607984), STUB1 (MIM: 607207), TAC3, TACR3, TUBB3 (MIM: 602661), and WDR11 (MIM: 606417). Of note, the case cohort in this study was not an unbiased cohort of individuals with IHH. Some individuals in the full database of individuals with IHH were excluded from this study because targeted Sanger sequencing revealed that they carry variants previously implicated in IHH (see above); thus, the cohort is partially depleted for variants in these genes. Also, some of the individuals in the cohort in this study were specifically included because they were known to be heterozygous for a variant in PROKR2; thus, the cohort is slightly enriched for PROKR2 mutations (see above).
To refine potential qualifying missense variants, missense variants in 20 genes chosen based on their strong evidence for causality in IHH were examined:9, 28 AXL, CHD7, FEZF1, FGF8, FGFR1, FGF17, GNRH1, GNRHR, HS6ST1, IL17RD, KISS1, KISS1R, NSMF, PROK2, PROKR2, SEMA3A, SOX10, TAC3, TACR3, and WDR11. These variants were annotated as described above, and variants with a MAF of less than 0.1% were used to assess how protein prediction algorithms could refine the performance of the burden testing algorithm.
Quantile-Quantile (QQ) Plot Metric
To measure inflation, we made an adaption to the genomic control (GC) metric commonly applied in GWAS.29 We name this metric . This adaptation allows for an assessment of inflation in summary statistics, while adjusting for the large number of genes with p value = 1.0 that results from the many genes with no variants observed in case cohort sequencing owing to relatively small case sample size. We calculated the observed and expected –log10(p values) at the 95th percentile of all genes. We then calculated the highest expected –log10(p value) among genes with p value = 1.0. is then calculated as below, where and are the observed and expected p values at the 95th percentile, and and are the observed and expected p values for the gene with the highest expected –log10(p value) among genes with p value = 1.0.
Software for Burden Testing
A software package for burden testing against public controls (TRAPD) is freely available (see Web Resources). The package is written in Python and R.
Results
Description of Approach
In the gene-based burden testing approach, the number of individuals carrying variants in a given gene is compared between disease case and control cohorts. Typically, sequencing data are filtered for rare, protein-altering variants, as these are believed to be more likely to cause a rare monogenic disorder.30, 31 These variants are referred to as “qualifying variants.”2 These filters reduce the noise that is introduced by benign variants. This exercise is repeated for each gene in the genome.
In this study, a gene-based burden test was performed for a cohort of 393 unrelated individuals with IHH against publicly available data as controls. The ascertainment and characteristics of these sequencing cohorts are described in Subjects and Methods. The case samples were all whole-exome sequenced across three separate sequencing platforms; variant-calling and quality control (QC) was performed jointly across the samples (see Subjects and Methods). The control samples were taken from the gnomAD database, which is comprised of 123,126 whole-exome sequencing samples.6 These samples were aggregated from many research projects assembled by many different centers and sequenced across several platforms; however, variant-calling and QC was performed jointly across the samples. Importantly, only summary-level data are available for gnomAD, rather than the full individual-level genotype data. As IHH is an extremely rare disorder (1 in 30–125,000),7, 8 gnomAD is likely to contain few if any individuals with IHH; therefore, gnomAD serves as a reasonable control dataset, particularly since prior work has demonstrated that for rare disorders, “contamination” of the control samples with individuals with the disorder has very limited impact on power to detect associated genes for rare disorders.1
In the following analyses, burden testing was performed to compare the case subjects (individuals with IHH) against a public control database (gnomAD) (Figure 1). To calibrate this approach, synonymous variants were used for comparison as they are presumably largely benign.30, 31 By utilizing synonymous variants from case sequencing as compared to gnomAD control subjects, we generated a “null” study to evaluate whether there might be artifactual inflation or deflation of test statistics as evaluated using a quantile-quantile (QQ) plot metric. Then, we systematically altered different thresholds and parameters to improve the quality and reliability of burden testing. Finally, we conducted burden testing on rare protein-altering qualifying variants to determine whether any of the 35 genes previously implicated in IHH could be “re-discovered” and if we could discover any genes that had not been previously associated with IHH.
Initial Burden Testing
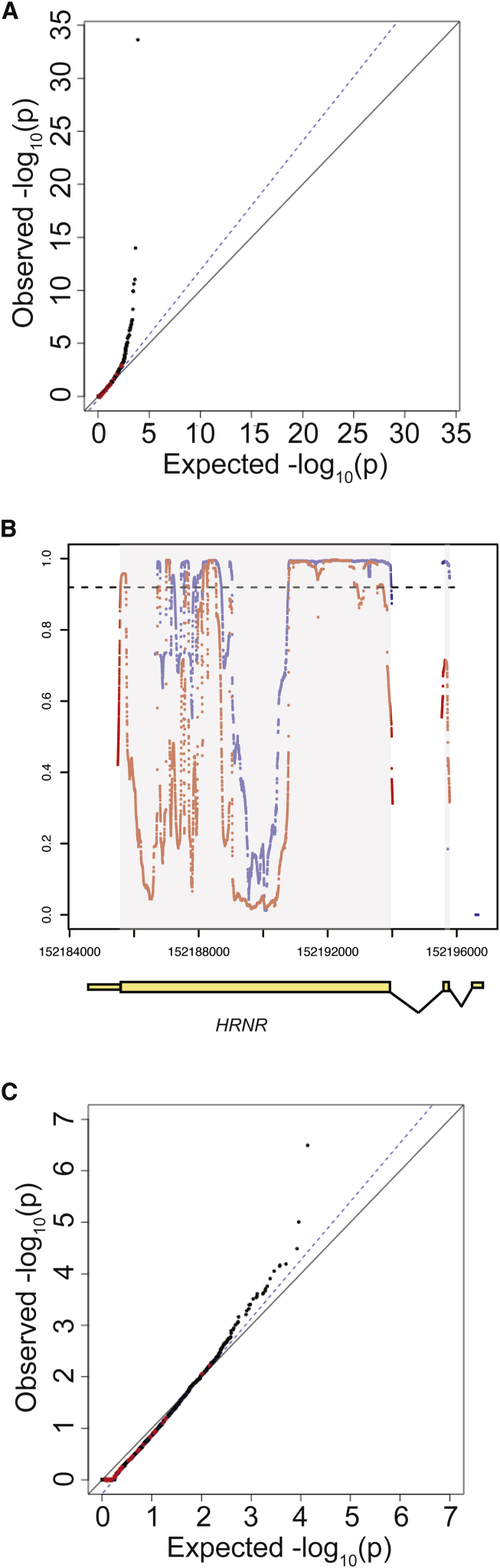
For the initial burden testing, the burden of rare (MAF < 0.1%), synonymous, single-nucleotide variants (SNVs) were compared between the case sequencing cohort and gnomAD control database without any filtering for variant quality or sequencing depth of coverage. As shown in Figure 2A, there was inflation of association statistics, with = 1.23, above the expected 1.00, suggesting the presence of artifact. Moreover, there were a large number of genes with association statistics that were more significant than expected (Figure 2A).
Figure 2.
Effect of Coverage on Distribution of Synonymous Variants
(A) Quantile-quantile plot of initial burden testing results using synonymous SNVs. Synonymous variants were used as they are likely mostly benign and can be used to test the null distribution. The x axis represents the expected –log10(p value) under the uniform distribution of p values. The y axis shows the observed –log10(p value) from the burden testing data. Each point is a single gene. Red dots represent the 35 genes previously implicated in IHH, while black dots represent the remaining genes in the genome. The black solid line shows the relationship between expected and observed p values under the uniform p value distribution. The dotted blue line shows the observed fit line between the 50th and 95th percentile of genes; the slope of this line is .
(B) Coverage at HRNR in case sequencing data and gnomAD control database. Exons are shown in yellow boxes below the plot, with wider boxes representing coding regions and narrower boxes representing UTRs. Introns (not drawn to scale) are shown as connecting lines between exons. Red dots represent coverage (as proportion of individuals with read depth >10×) in case cohort sequencing, while blue dots represent coverage in gnomAD control database. Each dot represents a single base. The dashed line represents the threshold for 90% of samples having sequencing read depth >10×.
(C) Repeat QQ plot from (A), except considering only bases for which more than 90% of samples had sequencing read depth >10× in both gnomAD and case sequencing data.
Effect of Read-Depth Filters
On closer examination, many genes appeared to exhibit differential read depth coverage between the case cohort and gnomAD controls. This difference likely reflects differences in exon-capture methods, resulting in differential coverage at targeted exons. For example, at HRNR (synonymous p value = 1.54 × 10−7), many bases were found to have much better sequencing coverage in case subjects as compared to control subjects (Figure 2B): 58.9% of coding bases in HRNR were covered at read depth of >10× in at least 90% of IHH case subjects, while 42.3% of the coding bases were covered at read depth of >10× in at least 90% of gnomAD control subjects (Figure 2B).
To overcome these striking differences in coverage, only sites that were covered at >10× in at least 90% of both case and control subjects were retained for analysis. After filtering these sites for depth of coverage, the association statistics demonstrated marked improvement (prior to read depth filtering: = 1.23, after read depth filtering: = 1.11, Figures 2A and 2C), suggesting that much of the inflation observed prior to read depth filtering was due to differences in sequencing coverage. For HRNR, following read depth filtering, the p value for synonymous variants was 0.48. However, there was still an excess of genes with inflated test statistics (Figure 2C), indicating that additional steps were needed to generate a well-calibrated analysis.
Effect of Quality Filters
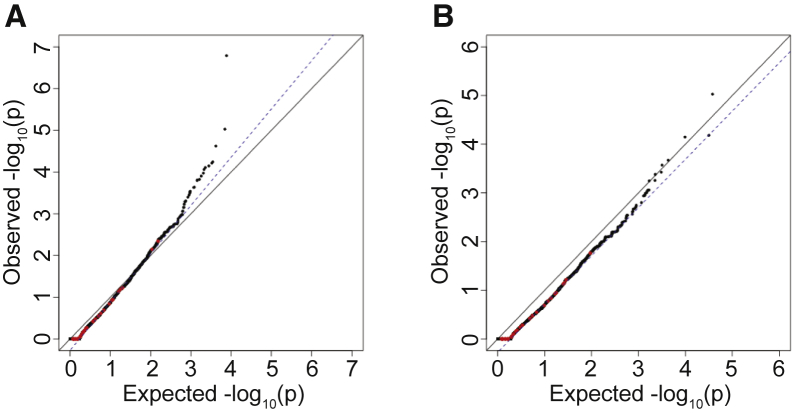
Even within sites well covered by the different sequencing platforms, some variants are called with higher confidence than others. Therefore, the results were further filtered to only those sites passing the quality thresholds of the GATK variant-calling software.14, 15, 16 Application of this additional filter removed many genes with p values that were more significant than expected but still resulted in inflated association statistics overall ( = 1.13, Figure 3A). On closer examination, many individual variants present in both the case sequencing cohort and gnomAD control database had discordant quality parameters, such that the same variant passed filters in one cohort but not the other. This probably occurred because of the vastly different sample sizes and possibly because of the incorporation of different samples when joint-calling each cohort. Among the 293,791 SNVs with MAF < 1.0% shared between the IHH cohort and gnomAD, 54,373 (18.5%) variants were discordant in their pass/fail status between the two cohorts. Thus, pass/fail status did not represent a consistent quality threshold across case and control sequencing data. To address this issue, other quantitative markers of quality were assessed.
Figure 3.
Effect of Variant Quality Filters on Distribution of Synonymous Variants
(A) Effect of adding pass/fail filters for variant quality. QQ plot of burden testing results following filtering for sites that passed GATK quality filters in the case and control sequencing data.
(B) Burden testing using QD scores to filter for sites. Only top 95% of sites in gnomAD based on QD scores and top 85% of sites in the case cohort sequencing based on QD scores are used.
Only sites where more than 90% of samples had sequencing read depth >10× in both gnomAD and the case cohort sequencing were considered (same as Figure 2B). QQ plots show burden testing results for synonymous variants.
Quantitative Measures of Variant Quality
To determine whether quantitative measures of quality might allow for improved quality filtering, two quality metrics output by the GATK variant-calling software were analyzed: variant quality score log-odds (VQSLOD) and quality-by-depth (QD).14, 15, 16 These scores have the advantage of being less dependent on the joint-calling procedure than the pass/fail filters set by GATK.16 Furthermore, as quantitative measures of quality, they allow greater flexibility in setting thresholds, as compared to binary pass/fail filters.
To evaluate the consistency of VQSLOD and QD as measures of quality, we examined the correlation in scores for rare (MAF < 1.0%) synonymous variants shared between the case and control sequencing data. Surprisingly, there was very little correlation for VQSLOD (r2 = 0.035) (Figure S1A), while QD displayed much better correlation (r2 = 0.61) (Figure S2A). Of note, at lower MAF bins, there was lower correlation in quality metrics between variants shared between the case sequencing cohort and gnomAD control database. This variable correlation at different MAFs reflects the importance of examining the performance of quality thresholds at the MAF of the intended comparison (Figures S1B, S1C, S2B, and S2C).
Given that QD was more strongly correlated at shared sites between the two cohorts at all MAFs, a QD filter was next applied to the data. We used a set of shared rare (MAF < 1%) SNVs to identify optimal QD score thresholds. These thresholds were identified separately for the case cohort sequencing and gnomAD control database. Subsequently, these QD score thresholds were applied to each cohort. Following experimentation with multiple combinations of filters, using the top 95% of sites in terms of QD scores for gnomAD and the top 85% of sites in terms of QD for the case sequencing cohort resulted in a well-controlled burden test (Figure 3B, = 0.99). This application of a percentile-based filter also allows for more adaptable thresholds; in contrast to a simple pass/fail filter, these thresholds are quantitative and can be set separately for the case and control sequencing data.
Analysis of Protein-Altering Variants
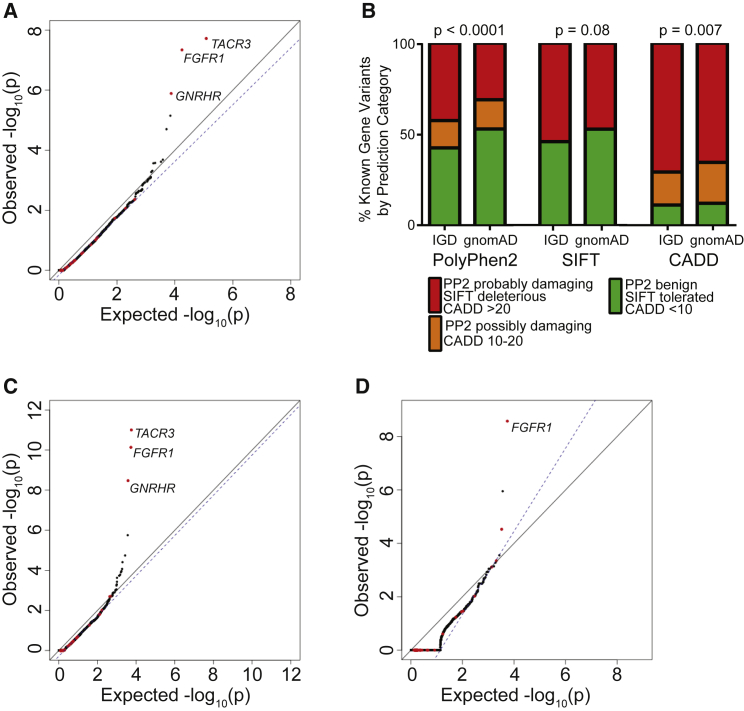
Having established a set of quality and read depth filters that appeared to generate a well-controlled test based on synonymous variants, these filters were used to analyze rare protein-altering qualifying variants, which are of interest. As an initial test, all SNVs that are predicted to alter protein sequence, including missense variants and protein-truncating variants (PTVs) (essential splice site, frameshift, and nonsense) were considered. When analyzing these protein-altering variants, the resulting QQ plot remained well controlled for artifact (Figure 4A, = 0.96). Additionally, many of the known IHH-associated genes emerged among the top genes, such as TACR3 (p value = 1.90 × 10−8, OR = 5.88), FGFR1 (p value = 4.56 × 10−8; OR = 4.92), and GNRHR (p value = 1.29 × 10−6, OR = 4.32). It is important to note that many of the genes previously implicated in IHH did not emerge as significant in the burden testing. This likely reflects the fact that many of these were identified in isolated families and contribute to only a small proportion of disease-affected case subjects.9 Also, as noted in Subjects and Methods, the cohort was partially depleted for individuals carrying variants in some genes associated with IHH.
Figure 4.
Selection of Damaging Variants to Improve the Power of Rare Variant Burden Testing
(A) Burden testing using all protein-altering variants.
(B) Distribution of PolyPhen2 (PP2), SIFT, and CADD scores among missense variants observed in IHH-affected case subjects as compared to gnomAD.
(C) Burden testing using only PTVs (essential splice site, frameshift, and nonsense) and missense variants computationally predicted to be damaging are considered.
(D) Burden testing using only PTVs.
For (A), (C), and (D), the same filters for coverage as in Figure 2B and variant quality as in Figure 3B were applied.
Calibrating with Protein Prediction Filters
Another reason for lack of significant associations for genes previously implicated in IHH is that many missense variants are benign and may limit the ability to detect enrichment of deleterious missense variants in case subjects as compared to control subjects. If so, computational predictions of the severity of biological effect of missense variants might improve the specificity of the gene-based burden test by removing noise introduced by benign missense mutations. Since there is no gold standard for prediction of pathogenic variants, the performance of three protein-prediction algorithms (PolyPhen2,20 SIFT,21 and CADD22) across missense variants in 20 genes implicated in IHH were examined, under the assumption that the IHH cohort would be enriched for damaging missense variants in these genes. These 20 genes, listed in Subjects and Methods, are a subset of the 35 genes previously implicated in IHH and were chosen based on their strong evidence of causality. To increase the number of observations, the analysis was expanded to include all missense changes in these 20 genes in 1,309 individuals with IHH for which Sanger sequencing of these genes had been performed. With these additional samples, there was significant enrichment in “probably damaging” score for PolyPhen2 (p < 0.0001). In contrast, SIFT demonstrated no significant enrichment, and CADD called more than 60% of all missense variants in individuals with IHH and gnomAD control subjecs as damaging (Figure 4B).
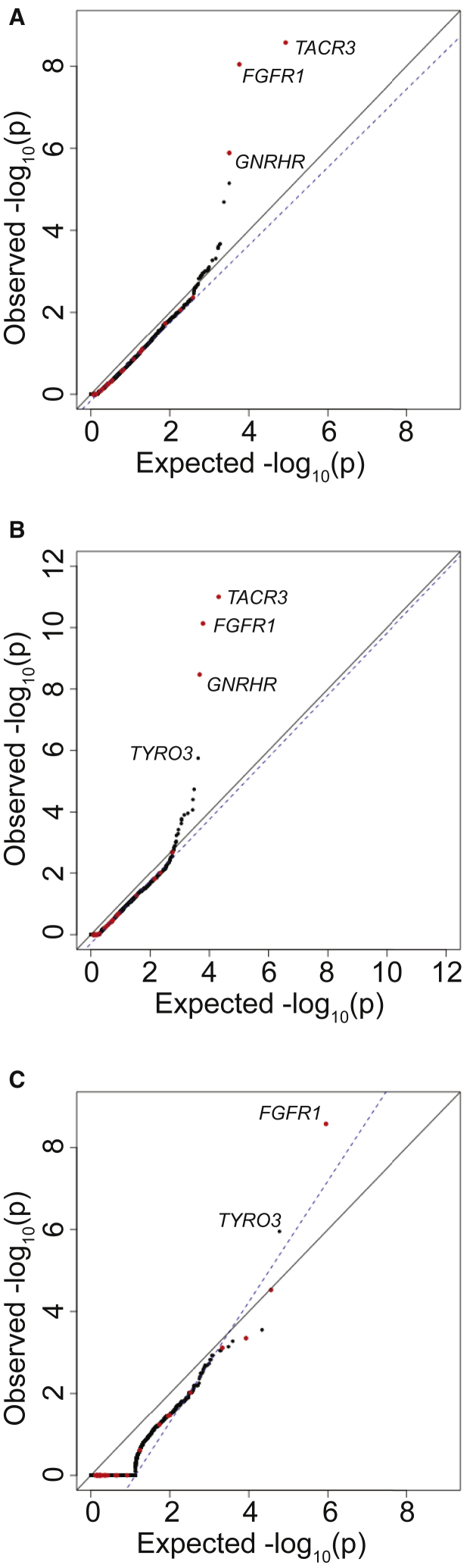
Since PolyPhen2 appeared to be the most consistent at distinguishing benign from pathogenic variants, subsequent burden testing was restricted to PTVs and missense SNVs scored as “probably damaging” by PolyPhen2 (Figure 4C, = 0.99). This additional filter resulted in improved test statistics for several genes previously implicated in IHH. For example, application of the PolyPhen2 filter improved the p value for FGFR1, a gene previously associated with IHH, from 4.56 × 10−8 (OR = 4.92) to 5.25−10 (OR = 7.01).
Finally, the performance of the burden test when using only protein-truncating SNVs (nonsense and essential splice site) was examined, as these are the most likely to be pathogenic and should in theory be the most enriched in the case cohort sequencing as compared to gnomAD control database. Previously associated genes such as FGFR1 (p value = 1.78 × 10−7; OR = 105.5) remained among the most highly associated genes. We note that although the association test statistics were not inflated on visual inspection, was 1.56 owing to the small number of genes carrying a PTV in the case cohort.
Addition of Indels to Burden Testing
Insertions and deletions (indels) typically result in greater rates of sequencing and variant-calling artifacts as compared to SNVs. However, adding indels in the top 75% in terms of QD scores in case cohort sequencing and the top 95% of indels for gnomAD samples continued to allow for a well-controlled burden test when testing all nonsynonymous sites (Figure 5A, = 0.95). When restricting to PTVs and missense variants nominated as “probably damaging” in PolyPhen2, additional incorporation of indels improved the association of FGFR1 from a p value of 5.25−10 to 7.31 × 10−11 (OR = 7.41; Figure 5B; = 0.979). Importantly, TYRO3, a candidate gene from mouse knockout studies,32, 33 achieved an exome-wide significance p value of 1.77 × 10−6 (OR = 7.98). Finally, when examining only protein-truncating variants, the addition of indels had minimal effects on the overall results as compared to burden testing without indels (Figure 5C): TYRO3 remained significant at p value = 1.11 × 10−6. These results demonstrate that indels, despite being common sources of artifact, can be incorporated into the burden testing and can improve power to detect disease genes.
Figure 5.
Addition of Indels to Rare Variant Burden Testing
For case cohort sequencing, SNVs in the top 85% of QD scores and indels in the top 75% were considered. For gnomAD, SNVs in the top 95% of QD scores and indels in the top 85% were considered. QQ plot shows burden testing using all nonsynonymous variants (A), PTVs (splice site, frameshift, and nonsense) plus missense variants computationally predicted to be damaging (B), or PTVs only (C).
Recessive Mode of Inheritance
As several genes implicated in IHH act in a recessive manner and the background rate of bi-allelic variants is low, testing variants under a recessive model might be a powerful way to detect disease genes. Therefore, a recessive test was constructed by tabulating the number of individuals with two qualifying variants in the case cohort and comparing it with the expected number of bi-allelic variant carriers in gnomAD. Since phase is unavailable in the case cohort sequencing, some individuals might actually have two variants on the same haplotype. Similarly, in the gnomAD control database, as individual-level data are not available, only an estimate of the number of individuals who would be bi-allelic could be calculated based on the frequency of rare heterozygous variants added to the number of homozygous variant carriers in each gene. Despite these limitations, performing the association under a recessive model produced strong association signals for many genes previously associated with IHH (Figure S3). Moreover, many genes that were not strongly associated under the dominant model reached much stronger association signals under the recessive model. For example, RNF216, a gene previously associated with IHH, reached a p value of 1.50 × 10−4 (OR = 204.5) under the recessive model, while it only reached a p value of 0.015 under the dominant model. In fact, mutations in RNF216 have been shown to cause disease only in a recessive state.34
Incorporation of Ancestry Data
Both the case and control cohorts are comprised of samples drawn from a variety of ancestries. Although the frequency of variants can differ vastly among individuals of different ancestries, the data still generated well-calibrated results using samples across all ancestries in both the case and control cohorts. Nonetheless, to assess the effect of ancestry on the results, we restricted the case and control subjects to individuals of European ancestry. While there is no way to directly compare the ancestries of the individuals in the case sequencing cohort with those in gnomAD, case cohort sequencing data were projected onto HapMap Phase 325 to identify those individuals who are of likely European ancestry (n = 263). For the gnomAD control database, the analysis was restricted to individuals listed as being of non-Finnish European ancestry (n = 55,860). The results were slightly deflated (Figure S4, = 0.95), and p values were less significant than when considering all ancestries, likely due to a loss of power from smaller sample sizes.
Results by Sequencing Batch
Given the fact that case cohort sequencing in this study was performed in three separate batches (see Subjects and Methods), we performed batch-specific burden testing. Similar to Figure 5B, qualifying variants were defined as variants with MAF < 0.1% and either PTVs or missense variants nominated as “probably damaging.” Overall, the burden testing results for each sequencing batch were well calibrated and many of the top genes remained consistent (Figures S5A–S5C). For one of the sequencing batches, there was some negative screening for samples carrying variants in genes previously associated with IHH, as well as some positive screening for individuals who are heterozygous carriers of PROKR2 variants. Consistent with the pre-screening, PROKR2 was among the strongest associations (p value = 2.5 × 10−3, OR = 9.60) (Figure S5B).
Using ExAC as a Control Cohort
Our approach can be applied to other public control databases. As a demonstration, we ran burden testing using an earlier exome aggregation database, ExAC, which contains 60,706 individuals (as compared to 123,136 in gnomAD).6 The overall results were similar from when using ExAC as a control cohort (Figure S6A, compare with Figure 5B), and that the top three genes remained as TACR3 (p value = 1.05 × 10−10), FGFR1 (p value = 1.44 × 10−9), and GNRHR (p value = 1.02 × 10−8). The overall correlation in p values was very strong when comparing results when gnomAD or ExAC was used as the control cohort (r2 = 0.90) (Figure S6B). However, genes previously associated with IHH tended to be more significant when using gnomAD as the control cohort (slope = 1.14 when comparing the –log10(p value) from when gnomAD was used as the controls as compared to when ExAC was used).
Additional Approaches for Correcting for Read Depth
In this paper, we chose to filter for only sites that are well covered in both case cohort sequencing and in gnomAD by filtering for sites where at least 90% of samples are covered at >10×. Previous papers have used similar approaches for adjusting for read depth. One approach that has been previously used is filtering for sites where there is no significant difference in the proportion of individuals covered.24 Application of this approach resulted in a similar number of coding bases analyzed as in our work (Figure S7A). Another approach that has been previously used is filtering for exons that are largely concordant in the number of bases covered in case as compared to control sequencing data.2 Application of this approach resulted in a much higher number of coding bases analyzed as compared to either of the first two approaches (Figure S7A). All three approaches for adjusting for read depth, when applied to our data, generated very similar results in burden testing (Figures 5B, S7B, and S7C).
Software Package for Burden Testing
To facilitate gene-based burden testing against public control databases, we created a freely available software package called TRAPD (Test Rare vAriants with Public Data). Users supply variant call data from whole-exome or whole-genome sequencing of disease cases. The software is readily able to compare case sequencing data to public exome-sequencing or genome-sequencing data from ExAC, gnomAD, or other control cohort where only summary-level data are available. This software allows for adaptable filtering on various quality and frequency fields to ensure a well-controlled burden test. The package is user friendly and requires minimal command line programming experience. It is also fast: burden testing can be completed in under 1 hr using just 4 GB of memory on a single node for 500 case samples against gnomAD.
Discussion
This study demonstrates that it is possible to generate a well-controlled and rigorous rare-variant gene-based burden test utilizing public control databases. The current study used whole-exome sequencing from 393 individuals with IHH. For controls, publicly available large-scale exome sequencing data from gnomAD (n = 123,126) was used. A gene-based burden testing strategy and software to perform burden testing against publicly available data was developed and tested sequentially to identify genes with enrichment of rare protein-altering variants in the case cohort. Rigorous sequential analyses progressively mitigated differences in sequencing platforms and variant calling between the case cohort sequencing and gnomAD control database. Ultimately, this methodology reliably “re-discovered” genes previously implicated in IHH and bolstered evidence for an existing candidate gene.
By applying iterative analyses, it was possible to discover and address various pitfalls encountered when using public control data. First, as different exome-capture and sequencing platforms were used between and within the case sequencing data and gnomAD control database, some regions of the genome were found to differ substantially in sequencing coverage. Since sequencing coverage has a strong impact on the ability to call variants, this resulted in artifactual associations (Figures 2A and 2B). Filtering for regions with high coverage eliminated artifact but also discarded many coding bases and occasionally entire genes. Other published approaches had similar overall results (Figure S7). Second, there were systematic differences in the quality metrics of variants between gnomAD and the case sequencing data, often resulting in highly discordant quality scores for the same variant between the datasets (Figures S1 and S2). This observation was likely related to the “data-dependent” nature of variant calling and the fact that the case data were called separately from the gnomAD database.6, 16 A critical insight is that traditional filtering for variants of high confidence, such as the use of pass/fail filtering, may not work well when there are systematic differences between case and control datasets that result from separate variant calling in the cohorts. This led to a key discovery that using quantitative measures of quality such as QD allow for highly adaptable fine-tuning of variant quality filters. QD is less dependent on the other samples present in the joint-calling set and can help mitigate the discordance that arises from comparing to a public control database.
This study also demonstrates that computational predictions of variant effect can impact results by filtering out statistical noise introduced by benign variants. As these filters are imperfect at distinguishing pathogenic from benign variants, they have an associated sensitivity and specificity which might affect power to detect certain genes. In this study, application of PolyPhen2 scores to the variant filtering improved the association statistics for some genes (Figures 4C and 5B). Furthermore, limiting analyses to PTVs (splice site, nonsense, and frameshift) also improves the association statistics for some genes (Figures 4D and 5C).
Interestingly, it appears that ancestry had a relatively small effect on results in this dataset, as using all samples across ancestries did not result in a skew of association statistics. Other groups have also observed well-calibrated results when performing burden testing in mixed ancestry cohorts,24, 35 likely because differences in burdens of rare protein-altering variants does not substantially differ across many ancestries, particularly in non-African populations that have not experienced severe population bottlenecks.6, 36, 37 Testing across all ancestries increases sample size and thus power and is a reasonable approach if the ancestries of case and control subjects are not highly discordant. The minimal effect of ancestry in our dataset may also be due to the relatively large and roughly comparable proportion of samples of European ancestry in the case sequencing cohort (66.9%) and gnomAD control database (45.3%), so may be less generalizable to case cohorts of other ancestries. Thus, it would be prudent for investigators to repeat their analyses before and after restricting the analyses to samples of the same ancestries within case and control subjects to ensure that their results are not affected by ancestry, especially when there is a mixture of samples with and without African ancestry or histories of population bottlenecks (e.g., Finnish). Additional caution may also be needed for admixed populations where case/control status is correlated with ancestry. A consideration for future methods development might be to test each ancestry individually and then perform a meta-analysis, or to adjust for ancestry through approaches such as principal components analysis.
This study demonstrates two of the characteristics that make certain genes more or less amenable to a burden testing strategy. First, locus heterogeneity can be a substantial barrier to burden testing.1 As expected, genes that contribute to a significant proportion of cases of IHH, such as FGFR1,38 are easily uncovered. However, many genes implicated in IHH contribute to only a small percent of cases of disease and were not uncovered by burden testing; often, no case subjects in our cohort carried variants in these genes. Second, autosomal-recessive gene RNF216 (n = 2 cases; p value = 1.50 × 10−4; OR = 204.5) can show statistical enrichment with a small number of case subjects.34 This is because the rate of protein-altering variants in these genes is so low in control subjects that just a few protein-altering variants observed in case subjects can be sufficient.
This approach can also expand the type of mutations associated with disease. Mutations in TACR3 acting in a bi-allelic manner have been shown to cause IHH in humans and further validated in the mouse model.39, 40 In fact, mice heterozygous for Tacr3 had no significant reproductive phenotype. Since this initial discovery, evaluation of humans with IHH has shown that heterozygous changes in TACR3, not just classic bi-allelic changes, may be capable of causing IHH.41 The findings in this paper provide statistical genetic evidence that heterozygous changes in TACR3 cause IHH. Similarly, GNRHR, which is classically autosomal recessive,42 demonstrated a statistical signal under a dominant model.
This type of large-scale analysis can bolster evidence for candidate genes, such as TYRO3, in this study. TYRO3 encodes the tyrosine-protein kinase receptor TYRO3, which is a part of the TYRO3-AXL-MER (TAM) receptor tyrosine kinase subfamily. Initially, this protein was implicated in hypogonadotropism in the setting of the loss of both Axl and Tyro3 in mice.32, 33 Notably, AXL has been previously implicated in IHH.43 In this study, protein-truncating mutations in TYRO3 were found to be enriched in individuals with IHH (p value = 1.77 × 10−6; OR = 7.98), providing evidence for the role of TYRO3 in IHH in humans. Additional validation of this finding in a separate cohort and functional studies of TYRO3 mutations will be needed to confirm TYRO3 as a bona fide gene associated with IHH.
In this work, we focused on gene discovery using public datasets as controls in a burden testing strategy. We are confident that our approach is effective given that we were able to “rediscover” genes previously associated with IHH. However, given our own lack of access to large control databases with individual-level genotyping data, we were unable to directly compare how well our approach compares with burden testing using individual-level control data. In the future, it would be interesting to evaluate how well burden testing performs when using individual-level control data versus summary-level public control data.
In summary, this study demonstrates how large, publicly available sequencing datasets can serve as controls in a gene-based burden testing strategy. Key sources of artifact were identified, and the effects of artifacts were successfully mitigated by calibrating burden testing using synonymous variants. This study demonstrates the need to systematically address potential sources of artifact, potentially by calibrating burden testing using synonymous variants, as is done herein, before reporting novel gene associations. Future reports of burden testing should provide evidence such as a genome-wide QQ plot and a metric such as to demonstrate that artifacts have been adequately addressed. This resource-efficient approach will allow researchers with rare disease cohorts but without access to large-scale control data to perform burden testing for discovery of genes associated with disease.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the affected individuals and their families for their participation in this study. We also thank the clinicians who have referred individuals over many years and members of the Massachusetts General Hospital Reproductive Endocrine Unit for discussions and reading of the manuscripts.
This work was supported by grant P50 HD28138 from the Eunice K. Shriver National Institute for Child Health and Human Development (NICHD). Y.-M.C. was supported by NIH R01 HD090071. J.N.H. was supported by NIH R01DK075787. M.F.L. was supported by a Catalyst Medical Research Investigator Training Award from Harvard Catalyst|The Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences, NIH award UL 1TR002541), and financial contributions from Harvard University and its affiliated academic healthcare centers. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, or the National Institutes of Health.
Published: September 27, 2018
Footnotes
Supplemental Data include seven figures and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.08.016.
Web Resources
Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/
ClinicalTrials.gov (identifier NCT00494169), https://clinicaltrials.gov
EIGENSOFT, https://data.broadinstitute.org/alkesgroup/EIGENSOFT/
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
International HapMap Project, ftp://ftp.ncbi.nlm.nih.gov/hapmap/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Samtools, http://www.htslib.org/doc/samtools.html
Segmental Duplications, https://github.com/ga4gh/benchmarking-tools/blob/master/resources/stratification-bed-files/SegmentalDuplications/mm-2-merged.bed.gz
Variant Effect Predictor, https://useast.ensembl.org/info/docs/tools/vep/index.html
Supplemental Data
References
- 1.Guo M.H., Dauber A., Lippincott M.F., Chan Y.-M., Salem R.M., Hirschhorn J.N. Determinants of power in gene-based burden testing for monogenic disorders. Am. J. Hum. Genet. 2016;99:527–539. doi: 10.1016/j.ajhg.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cirulli E.T., Lasseigne B.N., Petrovski S., Sapp P.C., Dion P.A., Leblond C.S., Couthouis J., Lu Y.-F., Wang Q., Krueger B.J. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S., Abecasis G.R., Boehnke M., Lin X. Rare-variant association analysis: study designs and statistical tests. Am. J. Hum. Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moutsianas L., Agarwala V., Fuchsberger C., Flannick J., Rivas M.A., Gaulton K.J., Albers P.K., McVean G., Boehnke M., Altshuler D., McCarthy M.I., GoT2D Consortium The power of gene-based rare variant methods to detect disease-associated variation and test hypotheses about complex disease. PLoS Genet. 2015;11:e1005165. doi: 10.1371/journal.pgen.1005165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B., Leal S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balasubramanian R., Crowley W.F., Jr. Isolated GnRH deficiency: a disease model serving as a unique prism into the systems biology of the GnRH neuronal network. Mol. Cell. Endocrinol. 2011;346:4–12. doi: 10.1016/j.mce.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laitinen E.-M., Vaaralahti K., Tommiska J., Eklund E., Tervaniemi M., Valanne L., Raivio T. Incidence, phenotypic features and molecular genetics of Kallmann syndrome in Finland. Orphanet J. Rare Dis. 2011;6:41. doi: 10.1186/1750-1172-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stamou M.I., Cox K.H., Crowley W.F., Jr. Discovering genes essential to the hypothalamic regulation of human reproduction using a human disease model: adjusting to life in the “-omics” era. Endocr. Rev. 2016;2016:4–22. doi: 10.1210/er.2015-1045.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Sykiotis G.P., Plummer L., Hughes V.A., Au M., Durrani S., Nayak-Young S., Dwyer A.A., Quinton R., Hall J.E., Gusella J.F. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc. Natl. Acad. Sci. USA. 2010;107:15140–15144. doi: 10.1073/pnas.1009622107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doty R.L., Shaman P., Dann M. Development of the University of Pennsylvania Smell Identification Test: a standardized microencapsulated test of olfactory function. Physiol. Behav. 1984;32:489–502. doi: 10.1016/0031-9384(84)90269-5. [DOI] [PubMed] [Google Scholar]
- 12.Dodé C., Teixeira L., Levilliers J., Fouveaut C., Bouchard P., Kottler M.-L., Lespinasse J., Lienhardt-Roussie A., Mathieu M., Moerman A. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinforma. 2013 doi: 10.1002/0471250953.bi1110s43. Published online October 15, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011;27:718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 22.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghavan N.S., Brickman A.M., Andrews H., Manly J.J., Schupf N., Lantigua R., Wolock C.J., Kamalakaran S., Petrovski S., Tosto G., Alzheimer’s Disease Sequencing Project Whole-exome sequencing in 20,197 persons for rare variants in Alzheimer’s disease. Ann. Clin. Transl. Neurol. 2018;5:832–842. doi: 10.1002/acn3.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Boehm U., Bouloux P.M., Dattani M.T., de Roux N., Dodé C., Dunkel L., Dwyer A.A., Giacobini P., Hardelin J.P., Juul A. Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism--pathogenesis, diagnosis and treatment. Nat. Rev. Endocrinol. 2015;11:547–564. doi: 10.1038/nrendo.2015.112. [DOI] [PubMed] [Google Scholar]
- 29.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 30.Goldstein D.B., Allen A., Keebler J., Margulies E.H., Petrou S., Petrovski S., Sunyaev S. Sequencing studies in human genetics: design and interpretation. Nat. Rev. Genet. 2013;14:460–470. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stitziel N.O., Kiezun A., Sunyaev S. Computational and statistical approaches to analyzing variants identified by exome sequencing. Genome Biol. 2011;12:227. doi: 10.1186/gb-2011-12-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierce A., Bliesner B., Xu M., Nielsen-Preiss S., Lemke G., Tobet S., Wierman M.E. Axl and Tyro3 modulate female reproduction by influencing gonadotropin-releasing hormone neuron survival and migration. Mol. Endocrinol. 2008;22:2481–2495. doi: 10.1210/me.2008-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce A., Xu M., Bliesner B., Liu Z., Richards J., Tobet S., Wierman M.E. Hypothalamic but not pituitary or ovarian defects underlie the reproductive abnormalities in Axl/Tyro3 null mice. Mol. Cell. Endocrinol. 2011;339:151–158. doi: 10.1016/j.mce.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margolin D.H., Kousi M., Chan Y.-M., Lim E.T., Schmahmann J.D., Hadjivassiliou M., Hall J.E., Adam I., Dwyer A., Plummer L. Ataxia, dementia, and hypogonadotropism caused by disordered ubiquitination. N. Engl. J. Med. 2013;368:1992–2003. doi: 10.1056/NEJMoa1215993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw N.D., Brand H., Kupchinsky Z.A., Bengani H., Plummer L., Jones T.I., Erdin S., Williamson K.A., Rainger J., Stortchevoi A. SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat. Genet. 2017;49:238–248. doi: 10.1038/ng.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Connor T.D., Kiezun A., Bamshad M., Rich S.S., Smith J.D., Turner E., Leal S.M., Akey J.M., NHLBIGO Exome Sequencing Project. ESP Population Genetics, Statistical Analysis Working Group Fine-scale patterns of population stratification confound rare variant association tests. PLoS ONE. 2013;8:e65834. doi: 10.1371/journal.pone.0065834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitteloud N., Meysing A., Quinton R., Acierno J.S., Jr., Dwyer A.A., Plummer L., Fliers E., Boepple P., Hayes F., Seminara S. Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol. Cell. Endocrinol. 2006;254-255:60–69. doi: 10.1016/j.mce.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 39.Topaloglu A.K., Reimann F., Guclu M., Yalin A.S., Kotan L.D., Porter K.M., Serin A., Mungan N.O., Cook J.R., Imamoglu S. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat. Genet. 2009;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J.J., Caligioni C.S., Chan Y.-M., Seminara S.B. Uncovering novel reproductive defects in neurokinin B receptor null mice: closing the gap between mice and men. Endocrinology. 2012;153:1498–1508. doi: 10.1210/en.2011-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gianetti E., Tusset C., Noel S.D., Au M.G., Dwyer A.A., Hughes V.A., Abreu A.P., Carroll J., Trarbach E., Silveira L.F.G. TAC3/TACR3 mutations reveal preferential activation of gonadotropin-releasing hormone release by neurokinin B in neonatal life followed by reversal in adulthood. J. Clin. Endocrinol. Metab. 2010;95:2857–2867. doi: 10.1210/jc.2009-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beranova M., Oliveira L.M.B., Bédécarrats G.Y., Schipani E., Vallejo M., Ammini A.C., Quintos J.B., Hall J.E., Martin K.A., Hayes F.J. Prevalence, phenotypic spectrum, and modes of inheritance of gonadotropin-releasing hormone receptor mutations in idiopathic hypogonadotropic hypogonadism. J. Clin. Endocrinol. Metab. 2001;86:1580–1588. doi: 10.1210/jcem.86.4.7395. [DOI] [PubMed] [Google Scholar]
- 43.Salian-Mehta S., Xu M., Knox A.J., Plummer L., Slavov D., Taylor M., Bevers S., Hodges R.S., Crowley W.F., Jr., Wierman M.E. Functional consequences of AXL sequence variants in hypogonadotropic hypogonadism. J. Clin. Endocrinol. Metab. 2014;99:1452–1460. doi: 10.1210/jc.2013-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.