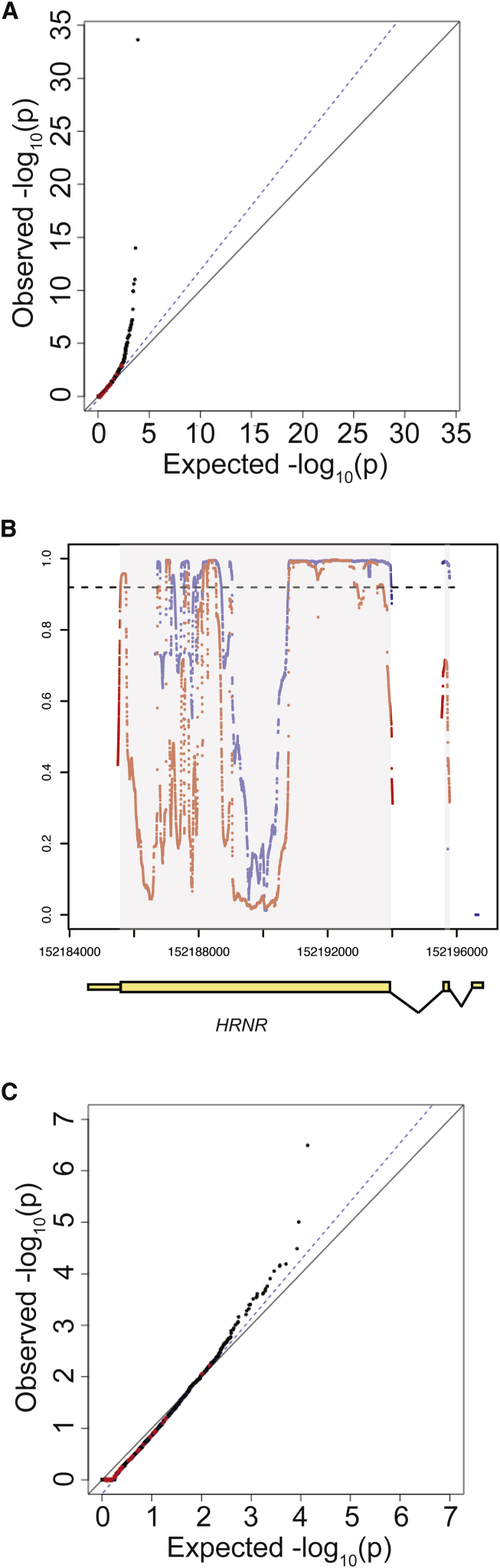
Figure 2.
Effect of Coverage on Distribution of Synonymous Variants
(A) Quantile-quantile plot of initial burden testing results using synonymous SNVs. Synonymous variants were used as they are likely mostly benign and can be used to test the null distribution. The x axis represents the expected –log10(p value) under the uniform distribution of p values. The y axis shows the observed –log10(p value) from the burden testing data. Each point is a single gene. Red dots represent the 35 genes previously implicated in IHH, while black dots represent the remaining genes in the genome. The black solid line shows the relationship between expected and observed p values under the uniform p value distribution. The dotted blue line shows the observed fit line between the 50th and 95th percentile of genes; the slope of this line is .
(B) Coverage at HRNR in case sequencing data and gnomAD control database. Exons are shown in yellow boxes below the plot, with wider boxes representing coding regions and narrower boxes representing UTRs. Introns (not drawn to scale) are shown as connecting lines between exons. Red dots represent coverage (as proportion of individuals with read depth >10×) in case cohort sequencing, while blue dots represent coverage in gnomAD control database. Each dot represents a single base. The dashed line represents the threshold for 90% of samples having sequencing read depth >10×.
(C) Repeat QQ plot from (A), except considering only bases for which more than 90% of samples had sequencing read depth >10× in both gnomAD and case sequencing data.