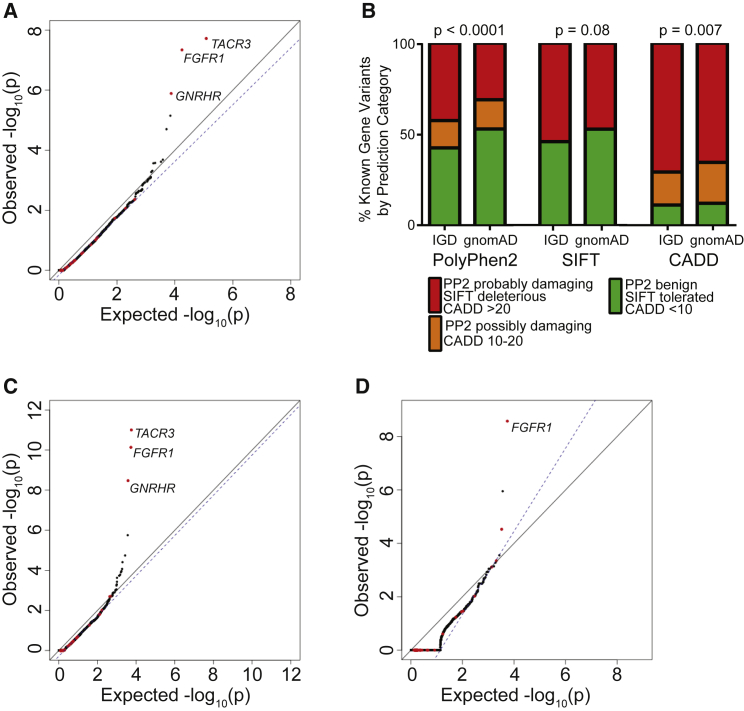
Figure 4.
Selection of Damaging Variants to Improve the Power of Rare Variant Burden Testing
(A) Burden testing using all protein-altering variants.
(B) Distribution of PolyPhen2 (PP2), SIFT, and CADD scores among missense variants observed in IHH-affected case subjects as compared to gnomAD.
(C) Burden testing using only PTVs (essential splice site, frameshift, and nonsense) and missense variants computationally predicted to be damaging are considered.
(D) Burden testing using only PTVs.
For (A), (C), and (D), the same filters for coverage as in Figure 2B and variant quality as in Figure 3B were applied.