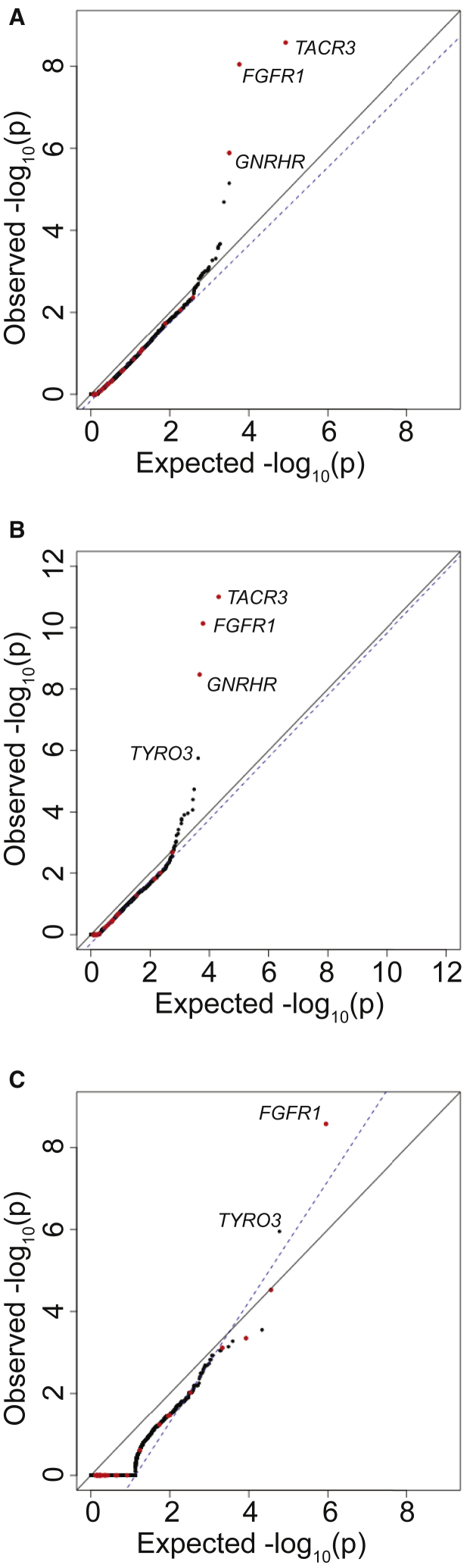
Figure 5.
Addition of Indels to Rare Variant Burden Testing
For case cohort sequencing, SNVs in the top 85% of QD scores and indels in the top 75% were considered. For gnomAD, SNVs in the top 95% of QD scores and indels in the top 85% were considered. QQ plot shows burden testing using all nonsynonymous variants (A), PTVs (splice site, frameshift, and nonsense) plus missense variants computationally predicted to be damaging (B), or PTVs only (C).