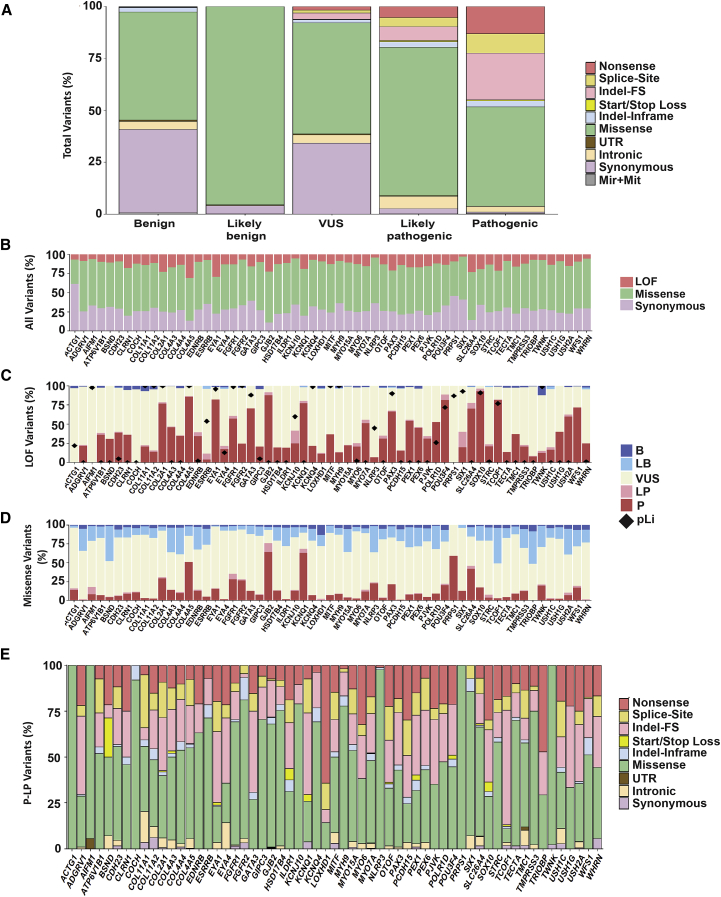
Figure 5.
Genomic Landscape of Deafness-Associated Genes
(A) Variant architecture by each classification category shows a strikingly distinct distribution of variant types across the five classifications.
(B) Distribution of LoF, missense, and synonymous variants is different across genes.
(C) Most LoF variants are P/LP and some genes are highly enriched in this type of variant.
(D) The contribution of missense variants to the mutational pool of hearing loss is variable across genes. However, in most genes, the majority of missense variants are VUSs.
(E) The mutational spectrum is gene specific. Splice-site indicates variants in canonical splice sites.
Only genes with ≥14 reported deafness-associated variants are included in this figure; the remaining genes are shown in Figures S4 and S5.