Figure 2.
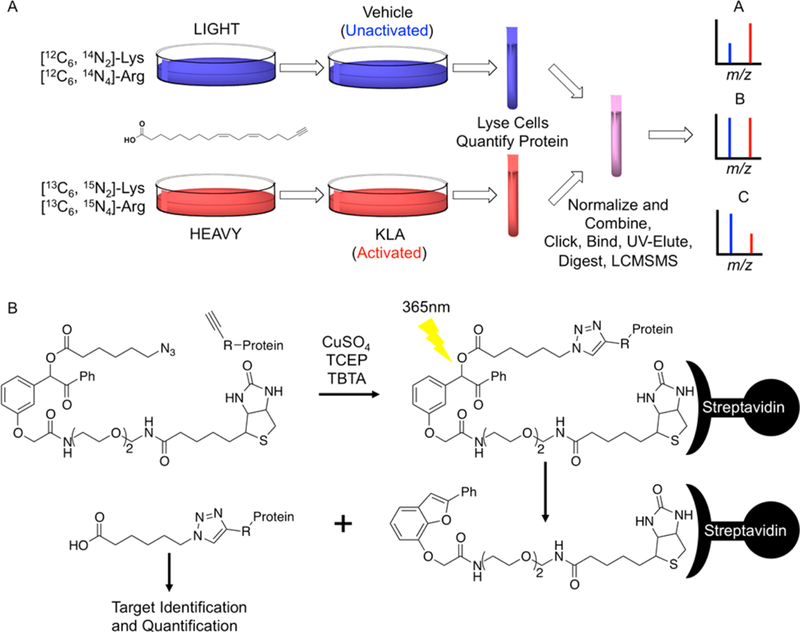
(A) SILAC workflow. Both heavy and light cell lines were incorporated with aLA. The light cell line was unactivated (vehicle control), while the heavy cell line was activated with KLA. After activation, the heavy and light cells were harvested and lysed, and the two samples were then combined in equal amounts of protein. Adducts were stabilized by reduction with NaBH4 and then attached to (B) UV-biotin by copper-mediated click chemistry for affinity enrichment of adducted proteins. MudPIT-MS/MS was used to analyze adducted proteins. (A) Three distinct protein classes can be observed that are described by different activated/unactivated ratios. Theoretical spectra for the three potential activated/unactivated ratios are seen for protein A where activated/unactivated > 1, protein B where activated/unactivated = 1, and protein C where activated/unactivated < 1.
