We have established a robust and reliable cell line-based glioma stem cell xenograft mouse model, genetically and epigenetically similar to its originating patient tumours. The survival of patients and injected mice correlate significantly, demonstrating that the transplanted mice mimic the clinical course of the patient.
Abstract
The leading cause of cancer-related mortality among children is brain tumour, and glioblastoma multiforme (GBM) has the worst prognosis. New treatments are urgently needed, but with few cases and clinical trials in children, pre-clinical models such as patient-derived tumour xenografts (PDTX) are important. To generate these, tumour tissue is transplanted into mice, but this yields highly variable results and requires serial passaging in mice, which is time-consuming and expensive. We therefore aimed to establish a cell line-based orthotopic mouse model representative of the patient tumour. Glioma stem cell (GSC) lines derived from paediatric GBM were orthotopically transplanted into immunodeficient mice. Overall survival data were collected and histological analysis of the resulting neoplasias was performed. Genome-wide DNA methylation arrays were used for methylation and copy-number alterations (CNA) profiling. All GSC lines initiated tumours on transplantation and the survival of the mice correlated well with the survival of the patients. Xenograft tumours presented histological hallmarks of GBM, and were also classified as GBM by methylation profiling. Each xenograft tumour clustered together with its respective injected GSC line and patient tumour based on the methylation data. We have established a robust and reproducible cell line-based xenograft paediatric GBM model. The xenograft tumours accurately reflected the patient tumours and mirrored the clinical course of the patient. This model can therefore be used to assess patient response in pre-clinical studies.
Introduction
Brain tumours are the most common solid tumour in children and paediatric high-grade gliomas, including glioblastoma multiforme (GBM), are among the most devastating forms with a 5-year survival of less than 20% (1). Recent studies using whole genome sequencing and DNA methylation analysis have shown that paediatric GBM differs from adult GBM in such ways that they cannot be viewed as the same disease (2,3). Point mutations in genes such as IDH and TP53, and methylated O6-methylguanine DNA methyltransferase (MGMT) are commonly detected in adult tumours but not in paediatric tumours. Instead, other mutations affecting the methylation pattern and chromatin structure have been observed, such as the hallmark missense mutations in the genes H3F3A and HIST1H3B (4). Paediatric tumours also harbour less copy-number alterations (CNA) than adult GBM, although some alterations occur. These include amplification of epidermal growth factor receptor (EGFR) and platelet-derived growth factor receptor alpha (PDGFRA), deletions of retinoblastoma 1 (RB1), phosphatase and tensin homolog (PTEN) and cyclin-dependent kinase inhibitor 2A/B (CDKN2A/B) (5–7). Even though collection of paediatric tumour tissue, through large collaborations like the Children’s Brain Tumour Tissue Consortium (8), allows for important studies of tumour cells, there are few in vitro and in vivo models. The majority of published in vitro models are cultured with serum (9), which does not retain the features of the tumours (10, 11), or in serum-free media as neurospheres (12, 13). Previously reported in vivo models on paediatric high-grade glioma include orthotopically injected tumour tissue, so called patient-derived tumour xenografts (PDTX) (14), and subcutaneously injected cell lines (12). However, reports of accumulating genetic alterations when culturing human tumour cells in mice due to a selection pressure from the host suggest that improvement of the PDTX model is needed (15). The transplantation of cell lines is more reliable and reproducible since the number of tumour-initiating cells is known; however, subcutaneous transplantations do not resemble the environment of the brain. So far, there are no well-described cell line-based orthotopic xenograft models for paediatric GBM. Such a model would be more effective than PDTX models as cells are not passaged in mice, thus reducing the number of animals needed as well as cost and time. We have recently published a study, describing the establishment and characterization of patient-derived glioma stem cell (GSC) cultures maintained under serum-free conditions (16). Here, we extended this study with thorough characterization of orthotopic cell line-based xenografts in immunocompromised mice initiated from these GSCs. The tumours that formed resembled the original tumours, and the disease course of the mice reflected the clinical course of the patients. This model is repeatable, reliable, stable and suitable for functional studies in the search of novel tumour markers and new treatments.
Material and methods
This study was carried out in accordance with guidelines and regulations approved by the regional ethics review board of Gothenburg (Dnr 604-12) and the Swedish Board for Agriculture (Dnr 10-2015). Signed informed consent was obtained from the children’s parents before tumour collection. Six paediatric GBM samples were collected at the Sahlgrenska University Hospital (Gothenburg, Sweden); all were primary tumours except for one relapsed tumour (GU-pBT-10). For patient data, see Table 1.
Table 1.
Patient data.
| Patient-ID | Gender | Age at diagnosis (years) | Location of the tumour | Primary or relapse | Outcome (days after diagnosis) |
|---|---|---|---|---|---|
| GU-pBT-7 | Male | 4.2 | Right hemisphere (thalamus) | Primary | DOD (325) |
| GU-pBT-10 | Male | 10.4 | Right hemisphere | Relapse | DOD (312) |
| GU-pBT-15 | Female | 12.5 | Brain stem | Primary | AWD (1238) |
| GU-pBT-19 | Male | 6.2 | Right hemisphere (thalamus) | Primary | DOD (485) |
| GU-pBT-23 | Female | 2.9 | Left hemisphere (temporal) | Primary | DOD (190) |
| GU-pBT-28 | Female | 11.1 | Pons (cerebellopontine angle) | Primary | DOD (237) |
AWD, alive with disease; DOD, dead of disease.
It should be noted that the mice in the study were untreated, whereas the patients received treatment. Treatment included neurosurgery, radiotherapy and chemotherapy in all cases, most commonly temozolomide in combination with bevacizumab, but other chemotherapy combinations were also used. The sole long-term survivor (GU-pBT-15), initially diagnosed as a primitive neuroectodermal tumour (CNS-PNET), received both focal and craniospinal irradiation followed by chemotherapy (mainly ifosfamide, carboplatin and etoposide) and valproic acid.
GSC cultures
Primary GSC cultures were established from the paediatric GBM samples collected at Sahlgrenska University Hospital during 2013–15 and have been previously described (16). Cell cultures were named accordingly: GU-pBT-7, GU-pBT-10, GU-pBT-15, GU-pBT-19, GU-pBT-23 and GU-pBT-28, and they refer to BPC-A7, BPC-B0, BPC-B5, BPC-B9, BPC-C3 and BPC-C8 in Wenger et al. (16). Cells were transplanted in passage 9–11 except for GU-pBT-23 transplanted in passage 19. The cell lines were tested and authenticated by short tandem repeat profiling (IdentiCell, Aarhus University Hospital) in 2016 and had no match to cell lines in DSMZ or ATCC cell banks nor to each other. The cell lines were also tested at regular intervals by single-nucleotide polymorphism profiling and compared with the primary tumour and earlier passages. All cell lines were tested negative for mycoplasma in higher passages by GATC Biotech.
Orthotopic transplantations
Immunocompromised 6- to 8-week-old female mice, with a body weight around 15 g (CIEA-NOG purchased from Taconic), were orthotopically xenotransplanted with human GSC lines, into the frontal cortex of the brain. 100 000 cells from six primary GSC cultures were injected into six groups of five mice each, using a stereotactic frame, as previously described (17). During the transplantation procedure, mice were anesthetized using isoflurane. Afterwards, they were monitored weekly for appearance and weight. On neurological symptoms and/or weight loss, mice were killed and their brains were immediately removed and fixed in 4% formaldehyde. Note that tissue/data come from the same cohort as used in Wenger et al. (16).
Immunohistochemistry of xenotransplanted mouse brain
Brains were embedded in paraffin and sectioned (5 µm) using a microtome. Sections were deparaffinized, heat-induced antigen unmasked (with citrate buffer pH 6.0) and stained according to Vecta Stain ABC kit (Vector) using rabbit monoclonal Nestin (human specific), ab105389 Abcam, 1:300; rabbit monoclonal Ki-67, ab92742 Abcam, 1:500; rabbit polyclonal glial fibrillary acidic protein (GFAP) Z 0334, Dako, 1:500. For histology, hematoxylin and eosin (HE) staining was used.
Genome-wide methylation arrays
DNA was extracted from patient tumours and formalin-fixed paraffin-embedded sections of xenotransplanted mice brains, and restored with REPLI-g (18) (only for formalin-fixed paraffin-embedded samples; 1500–2000 ng DNA), bisulfite-modified, and quality controlled as described previously (19), except that tissue was lysed for 48 h instead of 24 h during DNA extraction of formalin-fixed paraffin-embedded samples. DNA (1000 ng) from a mouse neural progenitor cell line was extracted using QIAamp® DNA micro kit (Qiagen, Hilden, Germany) and bisulfite-modified. According to the manufacturer’s instructions, 4 µl of bisulfite-modified DNA was processed on Infinium MethylationEPIC BeadChips (Illumina, San Diego, CA). Methylation data generated with HumanMethylation450BeadChips (Illumina) for the patient tumours and GSC lines comes from Wenger et al. (16).
Statistical methods
Methylation array data were analysed using R packages ChAMP and minfi. Probes for CpG sites on the X and Y chromosomes or close to a known single nucleotide polymorphism (20) were removed as well as probes present only on either HumanMethylation450BeadChips or Infinium MethylationEPIC BeadChips. In addition, probes that were detected using the Infinium MethylationEPIC BeadChip on DNA from the mouse neural progenitor cell line were removed, resulting in 250 438 probes that were kept for further analysis.
Hierarchical clustering was performed on probes with a standard deviation across samples above 0.25, using Euclidean distance metric and complete linkage. Differentially methylated probe analysis was performed on paired patient-xenograft differences, using differentially methylated probes with adjusted P-value <0.01 (Benjamini-Hochberg) and mean difference in β > 0.51. CNA were inferred from the methylation array data, using the R package conumee.
Results
The survival time of transplanted mice correlates significantly with the survival of the patients
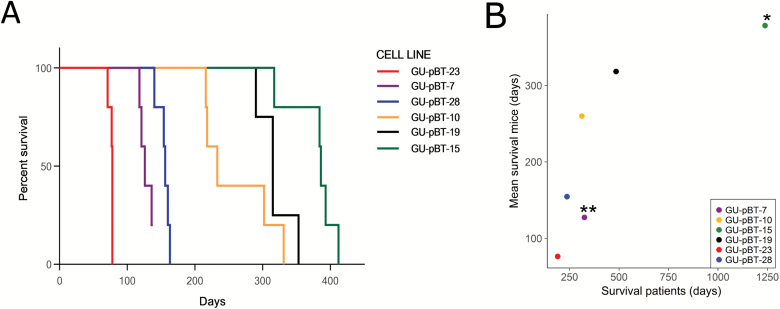
GSC lines derived from six patients (16) were orthotopically transplanted into immunocompromised mice. Survival time of patients and xenotransplanted mice was compared, and patient tumours, GSC lines and xenograft tumours were analysed for DNA methylation, CNA and classified with MethPed. Tumour take rate was close to 100% (a tumour was formed in 29/30 transplanted mice). The survival time for the mice in each group was close in time, as shown in the Kaplan-Meier curve (Figure 1A). The mean survival time of the mice in each group was significantly correlated to the survival of the patients, from whom the GSC cultures were established (Pearson correlation coefficient, r = 0.79, P-value = 3.5 × 10–7; Figure 1B).
Figure 1.
Cell-based xenotransplantation in mouse. (A) Kaplan-Meier curve shows that the survival within each group of GSC line injected mice are closely related in time. Tumour take-rate was high, with only 1 mouse of 30 not developing a tumour. (B) Mean survival of transplanted mice correlated with the survival of the patients (Pearson corr. = 0.79, P-value = 3.5 × 10–7). * = patients still alive, ** = one of five mice did not develop a tumour.
The morphology and methylation-based diagnosis of xenograft tumours resemble patient GBM
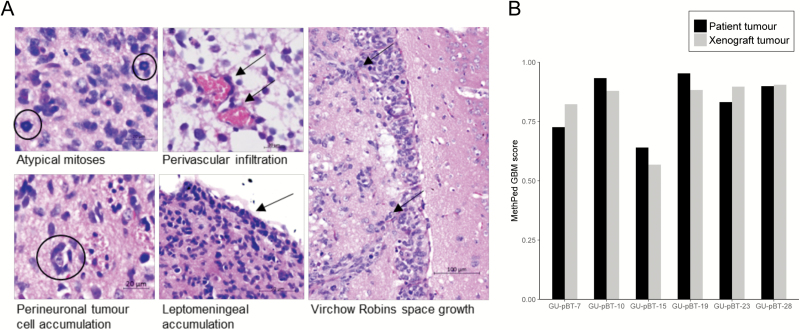
HE staining of patient tumours was performed at Sahlgrenska University Hospital and has partly been described (16). The tumour tissue exhibited characteristic GBM morphology with diffuse infiltration of anaplastic tumour cells showing high frequency of atypical mitoses and endothelial proliferation. The tumour cells frequently formed secondary structures of Scherer as perivascular infiltration, perineuronal and leptomeningeal accumulation. The tumours had variable expression of GFAP, strengthening the glial origin of the tumour, and a high number of proliferating cells (assessed by Ki-67). The samples were classified as GBM using the methylation-based classifier MethPed (16, 21). We next performed HE staining on the xenograft tumours. All GSC lines derived from patient GBM tumours gave rise to infiltrative tumours of human origin in immunocompromised mice (Figure 2). The xenograft tumour cells exhibited comparable growth patterns as the patient’s tumour cells with related morphology as described earlier (Figure 3A). The GSC culture GU-pBT-10 formed a focal tumour at the place of injection and showed limited infiltration of the surrounding brain parenchyma and less diffuse axonal growth. In contrast, the remaining GSC lines demonstrated a gliomatosis cerebri-like diffuse growth pattern. In the GU-pBT-19 xenograft, noticeable peritumoral oedema was present. Proliferation analysis using Ki-67 showed very high proliferation in all xenografts (Supplementary Figure 1). To additionally strengthen and confirm the diagnosis, we used our MethPed classifier (21), which diagnosed all xenografts as GBM with similar classification scores as the respective patient tumour (Figure 3B).
Figure 2.
Histology of xenotransplanted tumours. HE staining of the xenograft tumours displayed classical GBM growth pattern. Staining with a human-specific antibody against the neural stem cell marker Nestin verified the human origin of the tumours.
Figure 3.
Histology and methylation-based classification confirms GBM identity of xenograft tumours. (A) Enlargement of classical GBM growth pattern with asymmetric cell division; ring mitosis, structure of Scherer; axonal cell growth, perivascular infiltration and lepto-meningal accumulation of tumour cells. (B) MethPed classified the xenotransplanted tumours as GBM, with similar scores as for the patient tumours.
The methylome of the xenograft tumours is stable
Next, we performed hierarchical clustering on all β-values, measured with Illumina methylation BeadChips, with a standard deviation across samples above 0.25 (27 500 sites). Each group of samples, including patient tumour, GSC line and xenograft tumour, clustered together (Figure 4A). This indicates that the cells retain their characteristic methylation pattern in vitro and in vivo. Less than 3% of the studied CpG sites changed methylation status, defined as an absolute difference in β-value between samples above 0.51 (22), between either xenograft and patient tumour or cell line, strengthening the stability of the model (Figure 4B). We also looked for differentially methylated probes between paired xenograft and patient tumours. Only 127 CpG sites differed between the groups with a mean difference in β > 0.51, adjusted P-value <0.01.
Figure 4.
The methylome of the formed xenograft tumours is stable. (A) Hierarchical clustering of methylation β-values for 27 500 CpG sites, with standard deviation across samples above 0.25, groups the three samples (T – tumour, C – cell line and X – xenograft) from the same origin together for all cases. (B) Percentage of probes that change methylation status, defined as a difference in β-values >0.51 between xenograft and tumour (T) and cell line (C), respectively, was below 3% in all cases.
Key glioma genomic alterations are stable in the xenografts
We used the methylation array data to generate copy-number profiles for the samples (Supplementary Figure 2). In general, there was a good agreement between the patient tumour and the formed xenograft tumour, although some alterations were noted. EGFR and KRAS amplifications present in most patient tumours were also present in the corresponding xenograft tumours, as were deletions of the tumour suppressor genes RB1, PTEN and CDKN2A/B. Amplification on chromosome 4, where PDGFRA is located, was present in one of the patient tumours and in the corresponding xenograft tumour. Loss of chromosome 3 and 7 found in patient tumours was also identified in the corresponding xenografts. Some alterations were found de novo in the xenograft tumours. These were also detected in the GSC cultures that were transplanted, suggesting that these alterations were present already in the selected stem cell population.
Discussion
Recent efforts in profiling the genome and epigenome of paediatric brain tumours have led to DNA methylation-based classification and subgrouping of GBM tumours (3, 23). With new potential therapeutic targets, therapy can be designed more rationally, considering the diverse molecular features of these tumours (24). Genetically stable in vivo models are needed to pre-clinically test new drugs. In our cell line-based orthotopic xenograft model, we used well-characterized paediatric GSC lines, previously shown to be stable in culture; retaining the DNA methylome and CNA commonly found in the primary tumours (16). Here, we demonstrate that these GSCs are also stable in mice, initiating invasive tumours with a GBM-like growth pattern; anaplastic cells, atypic mitoses and secondary structures of Scherer. Tumour take-rate of engraftments is close to 100% and the cohort of mice injected with each cell line die within a very tight time frame, demonstrating accurate repeatability. This is in contrast to PDTX models, where a small portion of a heterogeneous tumour containing unknown numbers of cancer stem cells (25) results in large variabilities between animals and an uneven engraftment rate (26), or even failure of tumour growth (27). Xenograft tumours from PDTX models have also been shown to be influenced by the mouse microenvironment, rapidly acquiring CNA over multiple passages in animals as well as losing CNA present in the patient tumour (15). In comparison, although long-time cell cultures can accumulate genetic alterations, key glioma CNA found in patients were retained in our cell line-based mouse model, and the xenograft tumours presented a stable methylome with similar methylation pattern as the patient tumour. Less effort in terms of animal care and surgical procedures are needed and the number of animals is reduced, which is preferable both from an ethical and economical point of view.
Importantly, this thorough characterization of the xenograft tumours and strong correlation to the patient survival add a dimension of clinical usefulness to previous published work on our cell culture system. This is, to the best of our knowledge, the first paediatric GBM model that mimics the clinical course of the patient. It further enables studies to expand our understanding of the pathology and molecular driving forces behind paediatric GBM; an important step towards finding new treatments and improving the prognosis for affected children.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Funding
This work was supported by the Swedish Cancer Society, the Swedish Children’s Cancer Society, the Swedish Research Council, the Swedish Society for Medical Research, the Wenner-Gren foundation, Wilhelm and Martina Lundgren, Assar Gabrielsson, The Royal Physiographic Society of Lund (the Nilsson-Ehle Endowments), Sahlgrenska Universitetssjukhusets fonder and a Marie Curie CIG from the EU’s Seventh Framework Programme (FP7).
Acknowledgements
We thank Malin Hagberg Thulin for technical assistance and UCL Genomics for DNA methylation array processing.
Conflict of Interest Statement: None declared.
Abbreviations
- CNA
copy-number alterations
- GBM
glioblastoma multiforme
- GSC
glioma stem cell
- HE
hematoxylin and eosin
- PDTX
patient-derived tumour xenografts
References
- 1. Broniscer A. (2006)Past, present, and future strategies in the treatment of high-grade glioma in children. Cancer Invest., 24, 77–81. [DOI] [PubMed] [Google Scholar]
- 2. Sturm D., et al. (2014)Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat. Rev. Cancer, 14, 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones C., et al. (2014)Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat. Rev. Cancer, 14, 651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Venneti S., et al. (2014)A sensitive and specific histopathologic prognostic marker for H3F3A K27M mutant pediatric glioblastomas. Acta Neuropathol., 128, 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sturm D., et al. (2012)Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell, 22, 425–437. [DOI] [PubMed] [Google Scholar]
- 6. Wu G., et al. (2012)Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet., 44, 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paugh B.S., et al. (2010)Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J. Clin. Oncol., 28, 3061–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Children’s Brain Tumor Tissue Consortium(2017).https://cbttc.org/(30 November 2017, date last accessed).
- 9. Xu J., et al. (2015)Pediatric brain tumor cell lines. J. Cell. Biochem., 116, 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee J., et al. (2006)Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell, 9, 391–403. [DOI] [PubMed] [Google Scholar]
- 11. Li A., et al. (2008)Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol. Cancer Res., 6, 21–30. [DOI] [PubMed] [Google Scholar]
- 12. Josupeit R., et al. (2016)Pediatric and adult high-grade glioma stem cell culture models are permissive to lytic infection with parvovirus H-1. Viruses, 2016, 8, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu J., et al. (2012)Novel cell lines established from pediatric brain tumors. J. Neurooncol., 107, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Houghton P.J., et al. (2007)The pediatric preclinical testing program: description of models and early testing results. Pediatr. Blood Cancer, 49, 928–940. [DOI] [PubMed] [Google Scholar]
- 15. Ben-David U., et al. (2017)Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet., 49, 1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wenger A., et al. (2017)Stem cell cultures derived from pediatric brain tumors accurately model the originating tumors. Oncotarget, 8, 18626–18639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pollard S.M., et al. (2009)Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell, 4, 568–580. [DOI] [PubMed] [Google Scholar]
- 18. Thirlwell C., et al. (2010)Genome-wide DNA methylation analysis of archival formalin-fixed paraffin-embedded tissue using the Illumina Infinium HumanMethylation27 BeadChip. Methods, 52, 248–254. [DOI] [PubMed] [Google Scholar]
- 19. Kling T., et al. (2017)Validation of the MethylationEPIC BeadChip for fresh-frozen and formalin-fixed paraffin-embedded tumours. Clin. Epigenetics, 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nordlund J., et al. (2013)Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol., 14, r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Danielsson A., et al. (2015)MethPed: a DNA methylation classifier tool for the identification of pediatric brain tumor subtypes. Clin. Epigenetics, 7, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guilhamon P., et al. (2014)Assessment of patient-derived tumour xenografts (PDXs) as a discovery tool for cancer epigenomics. Genome Med., 6, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwartzentruber J., et al. (2012)Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature, 482, 226–231. [DOI] [PubMed] [Google Scholar]
- 24. McCabe M.T., et al. (2014)EZH2 as a potential target in cancer therapy. Epigenomics, 6, 341–351. [DOI] [PubMed] [Google Scholar]
- 25. Singh S.K., et al. (2004)Identification of human brain tumour initiating cells. Nature, 432, 396–401. [DOI] [PubMed] [Google Scholar]
- 26. Shu Q., et al. (2008)Direct orthotopic transplantation of fresh surgical specimen preserves CD133+ tumor cells in clinically relevant mouse models of medulloblastoma and glioma. Stem Cells, 26, 1414–1424. [DOI] [PubMed] [Google Scholar]
- 27. Kim K.M., et al. (2016)Failure of a patient-derived xenograft for brain tumor model prepared by implantation of tissue fragments. Cancer Cell Int., 16, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.