Abstract
To evaluate the contribution of association studies of candidate polymorphisms to inherited predisposition to Hodgkin lymphoma (HL), we conducted a systematic review and meta-analysis of published case-control studies. Of the variants examined more than once in candidate gene association studies, we identified 21 studies that reported on 12 polymorphic variants in 10 genes. Data were also extracted from a published genome wide-association study to allow analysis of an additional 47 variants in a further 30 genes. Promising associations were seen in nine of the variants (p < 0.05). Given that the estimated false positive report probabilities (FPRPs) for all associations are high (i.e. FPRP > 0.2), these findings should be interpreted with caution. While studies of candidate polymorphisms may be an attractive means of identifying risk factors for HL, future studies should employs ample sizes adequately powered to identify variants having only modest effects on HL risk. Furthermore, because of aetiological heterogeneity within HL, stratification of genotyping according to age, tumour Epstein-Barr virus status and histology is essential.
Keywords: Hodgkin Lymphoma, Genetic susceptibility, Meta-analysis, Systematic review, Candidate gene, Polymorphism
Introduction
Hodgkin Lymphoma (HL) is a B-cell malignancy affecting ~3 per 100,000 of the population each year in most Western countries(1). HL is typically classified based on histopathological appearances into classical HL (cHL) which accounts for 95% of cases and nodular lymphocyte predominant HL (NLPHL). The presence of EBV latent membrane protein and/or EBV-encoded small RNAs (EBERs) in Hodgkin Reed-Sternberg cells (HRS) defines EBV-positive disease(2).
Hodgkin lymphoma shows a bimodal age distribution in incidence, with geographic specific differences. In economically developed countries, the first peak of incidence typically occurs around 20 years of age with a second peak around 65 years of age(1). Evidence for the existence of inherited genetic predisposition to HL is derived from the high concordance rates in identical twins(3) and from case-control studies and cohort studies which have shown a 3-fold increased risk of HL in relatives of HL patients(4).
The failure to identify a major disease-causing locus has led to the proposal that a significant component of the inherited susceptibility is enshrined in the co-inheritance of multiple risk alleles, some of which are likely to be common. The most frequent method for identifying prevalent low risk variants is through association studies based on comparing the frequency of polymorphic genotypes in cases and controls. Alleles positively associated with the disease are analogous to risk factors in epidemiology and may be causally related to disease risk or be in linkage disequilibrium (LD) with disease-causing variants. There are a number of different methods of analysing the risk associated with a specific variant. For simple bi-allelic polymorphisms, the odds ratio (OR) of disease can be derived by comparing allele frequencies in cases and controls. Alternatively, a comparison of frequencies of the three genotypes among cases and controls can be made, using homozygosity of the “wild-type allele” as the reference group. Where homozygotes are rare, heterozygotes and homozygotes are grouped together, but this is only appropriate if a dominant model of disease susceptibility can be assumed. Similarly, combining heterozygotes with wild-type homozygotes is only appropriate if alleles act recessively.
The genetic candidates that have been evaluated as susceptibility genes for HL to date can be divided into the following groups: immune function/response, carcinogen metabolism enzymes, folate metabolism enzymes, DNA repair proteins, and others (table 1). We elected to exclude candidate gene association studies exploring the human leucocyte antigen (HLA) region and HL given the already established predisposition at the HLA region(5) and the complex LD structure making identification of the disease-causing variant problematic.
Table 1. Polymorphisms studied as risk factors for Hodgkin Lymphoma.
| Class/Gene | Polymorphism | Type | AA change | Effect | Methods of detection |
|---|---|---|---|---|---|
| Immune response | |||||
| TNFα | rs1800629 A>G | Upstream | None | Reduced expression | Taqman |
| rs1800750 G>A | Upstream | None | Reduced expression | Taqman | |
| FCGR2 | rs1801274 C>T | Missense | A519C, R131H | Altered activity | Pyrosequencing |
| IL1A | rs1800587 C>T | Upstream | None | Increased expression | Taqman |
| IL1B | rs16944 G>A | Upstream | None | Altered expression | Taqman |
| IL4 | rs2243248 T>G | Upstream | None | Altered expression | Cycling temperature denaturing electrophoresis |
| rs2243250C>T | Upstream | None | Altered expression | Sequenom MassARRAY iPLEX | |
| IL4A | rs1801275 A>G | Missense | Q551R | Altered activity | Sequenom MassARRAY iPLEX |
| IL6 | rs1800795 G>C | Upstream | None | Reduced expression | Taqman |
| IL10 | rs1800890: T>A | Upstream | None | Lower expression? | Cycling temperature denaturing electrophoresis, PCR |
| rs1800896 G>A | Upstream | None | Lower expression> | RFLP Taqman, SSCP | |
| IL10RA | rs2229113: A>G | Missense | G330R | Altered activity | PCR-RFLP |
| rs3135932 A>G | Missense | S159G | Altered activity | PCR-RFLP | |
| IRF4A | rs872071 A>G | Upstream | None | Altered expression | Allele-Specific PCR |
| LTC4S | rs730012 A>C | Upstream | None | Altered expression | Taqman |
| CXCL12 | rs1801157 G>A | Upstream | None | Altered expression | PCR-RFLP |
| TLR1 | rs5743551 A>G | Upstream | None | Increased expression | PCR-RFLP |
| TLR2 | rs4696480 T>A | Upstream | None | Altered expression | PCR-RFLP |
| TLR9 | rs187084 T>C | Upstream | None | Altered expression | PCR-RFLP |
| rs5743836 T>C | Upstream | None | Altered expression | PCR-RFLP | |
| Carcinogen metabolism | |||||
| GSTM1 | Deletion | NA | NA | Abolishes activity | PCR-RFLP |
| GSTT1 | Deletion | NA | NA | Abolishes activity | PCR-RFLP |
| GSTP1 | rs1695 A>G | Missense | I105V | Decreased activity | Cycling temperature denaturing electrophoresis, PCR-RFLP |
| GSTA1 | rs3957357 C>T | Upstream | None | Altered expression | Cycling temperature denaturing electrophoresis |
| EPHX | rs1051740 T>C | Missense | T113C | Altered activity | PCR-RFLP |
| rs2234922 A>G | Missense | H139G | Altered activity | PCR-RFLP | |
| CYP2C9 | rs1057910 A>C | Missense | I359L | Altered activity | Taqman |
| rs1799853 C>T | Missense | R144C | Altered activity | Taqman | |
| DNA repair | |||||
| XPA | rs1800975 G>A | Missense | A23G | Altered activity | Taqman |
| XPC | rs2228000 A>G | Missense | A499V | Altered activity | Sequenom MassARRAY IPLEX |
| rs2228001 A>G | Missense | K940Q | Altered activity | Taqman | |
| XPF/ERCC1 | rs3212986 C>A | Missense | Q504K | Altered activity | Taqman |
| XPG/ERCC5 | rs17655 G>C | Missense | D1104H | Altered activity | Taqman |
| XRCC1 | rs1799782 C>T | Missense | R194W | Altered activity | Sequenom MassARRAY iPLEX |
| XRCC3 | rs861539 C>T | Missense | T241M | Altered activity | Taqman |
| Folate Metabolism | |||||
| MTHFR | rs1801133 C>T | Missense | A222V | Decreased activity | Taqman, Melting curve analysis, PCR-RFLP |
| MTR | rs1805087 A>G | Missense | D919G | Reduced activity | Taqman |
| Others | |||||
| COX2 | rs5277 C>G | Missense | G102C | Altered activity | Taqman |
| rs20417 G>C | Upstream | None | Decreased activity | Sequenom MassARRAY iPLEX, Taqman, Pyrosequencing | |
| rs689466 G>A | Upstream | None | Decreased activity | ||
| ABCC2 | rs17222723 T>A | Missense | V1188E | Altered activity | Cycling temperature denaturing electrophoresis |
| NBN | rs1801282 C>A | Missense | P12A | Altered activity | Sequenom MassARRAY iPLEX |
| TP53 | rs1042522 | ||||
| NFKB1 | rs3774937 | ||||
| NFKB1A | rs696 | ||||
| rs8904 | |||||
| rs1050851 | |||||
| rs19571006 | |||||
| CHUK | rs2230804 | ||||
| PTGES | rs10448290 | ||||
| rs2241270 | |||||
| rs4837404 | |||||
| HPSE | rs4693608 | ||||
| rs11099592 | |||||
| rs436425 | |||||
| UGT1A1 | NA | Tandem Repeats | None | Reduced promoter activity | PCR-RFLP, Sanger sequencing |
AA, amino acid; NA, not applicable; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism;
Although some polymorphic variants have only been examined once, most have been evaluated as risk factors in several studies but often with discordant findings. Furthermore, many of the studies have been based on small sample sizes with limited power to robustly demonstrate a relationship with HL risk. To gain better insight into the impact of polymorphic variants on risk of HL, we have undertaken a systematic review of published studies and used standard meta-analysis techniques according to Cochrane(6) In addition, we have used genotype data from a published Genome-Wide Association Study (GWAS) to perform a meta-analysis on variants that have only been examined once in the literature (7, 8).
Methods
Study identification
A literature search for studies reporting on the association between polymorphic variants and HL was conducted using the electronic database PubMed from May 1991 - December 2014. The search strategy included using the keywords ‘Hodgkin Lymphoma’, ‘case’, ‘control’, ‘polymorphism’, ‘risk’, ‘genetic, association’. We searched for additional studies in the bibliographies of identified publications, including previous review articles and meta-analyses(9, 10).
Selection criteria
Studies were eligible if they were based on unrelated individuals and examined the association between HL and variants in candidate genes chosen based on a priori knowledge of HL/cancer biology. Variants were only carried forward for meta-analysis if full genotype data for both cases and controls were extractable from the article. Only studies published as full-length articles or letters in peer-reviewed journals in English were included in the analysis.
Data extraction
Data for analyses, including study design, sample size and ethnicity as well as allele and genotype frequencies, were extracted from the published articles and summarised in a consistent manner to aid comparison.
Data extraction from Genome Wide Association Studies
Where possible, genotype data from 1,465 cases and 6,417 controls were extracted from published GWAS to allow the inclusion of variants examined once in the literature (7, 8).
Statistical analysis
Raw data for genotype frequencies, without adjustment, were used for calculation of the study-specific estimates of odds ratio and 95% confidence limits (CIs). Cochran’s Q-statistic was used to test for heterogeneity, and the percentage variability of the pooled OR attributable to heterogeneity between studies was quantified using the I2 statistic (large heterogeneity typically defined by I2>75%). A p-value>0.05 for the Q test was considered to indicate a lack of heterogeneity across studies, so the pooled estimation of the OR of each study was calculated by the fixed-effects model (11). Otherwise, the random effects model was employed(12). The significance of the pooled OR was determined by the Z-test and p<0.05 was considered as statistically significant. An estimate of the potential publication bias was conducted by the examination of funnel plots, in which the standard error (SE) of log(OR) of each study was plotted against its log(OR). An asymmetric plot is reflective of publication bias. The funnel plot symmetry was assessed by Egger’s test based on inverse-variance weighted regression of the effect sizes on their precision (the inverse of standard error), to test whether the intercept deviated significantly from zero; a p<0.05 was considered indicative of statistically significant publication bias(13). To test for population stratification (i.e. deviation from Hardy-Weinberg equilibrium (HWE), the distribution of genotypes in control subjects of each individual population was tested for departure by means of the χ2 test(14). For each statistically significant association identified, we estimated the false positive reporting probability (FPRP) (15). The FPRP value is determined by the p-value, the prior probability for the association, and statistical power. We calculated FPRP assuming a prior probability of 0.001 proposed for candidate gene analyses(16). Statistical power was based on the ability to detect an OR of 1.2 and 1.5 (or reciprocal), with α equal to the observed p-value. To evaluate whether the association was noteworthy, we imposed a FPRP cut-off value of 0.2, advocated for summary analyses. Hence, FPRP values < 0.2 were considered to indicate robust associations (15).
Statistical analyses were undertaken using Stata version 10.0 (Stata Corporation, College Station, TX).
Results
Characteristics of published studies
Meta-analysis of variants studies more than once in the literature
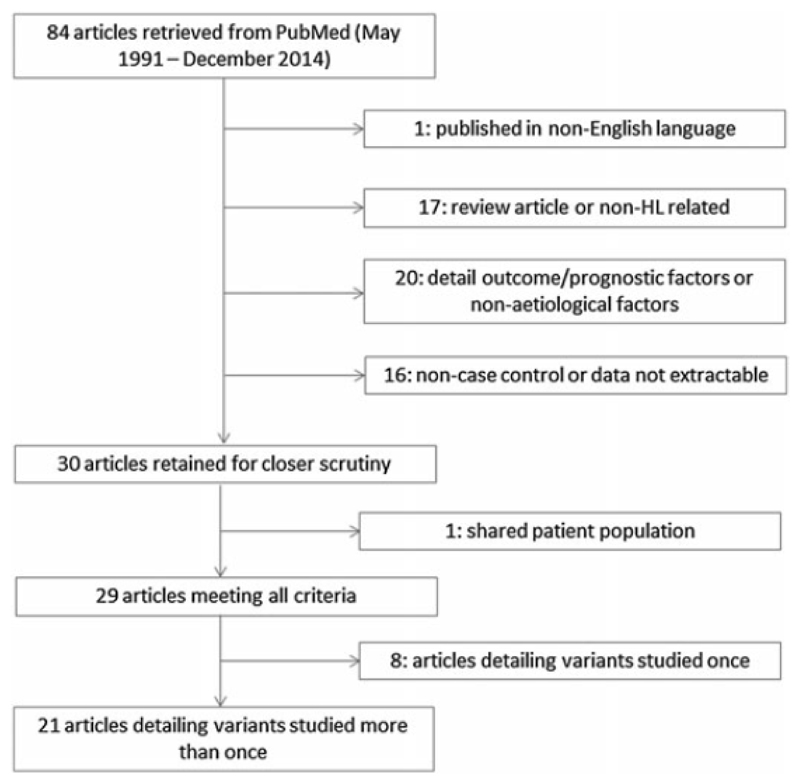
We retrieved 84 published studies using our search criteria (Figure 1). Of variants examined more than once in the literature we identified a total of 21 publications detailing 12 variants in 10 genes (Table 1). Six of the variants were genotyped in the published GWAS. All of the published the studies were essentially of similar design, although different types of controls have been analysed (Table 2). In the final group, six studies included data on ethnicity of cases and controls (17–21), three studies included data on EBV-status (17, 22, 23), and eight studies included data on histopathological subtype of HL (17, 19, 20, 22, 24–27). Two studies stratified genotype by histology (23, 25), and two studies stratified genotypes by EBV-status (22, 23). ORs of HL associated with each polymorphism in individual studies are detailed in the Table 3.
Figure 1.
Inclusion and exclusion criteria for studies
Table 2. Studies of polymorphisms and risk of Hodgkin lymphoma.
| Reference | Place of study | Gene studied | Ethnicity | N | Cases | N | Controls |
|---|---|---|---|---|---|---|---|
| Ruiz-Cosano J et al, 2013 | Spain |
XPG.ERCC5 MTR |
Caucasian | 20 | HL a subset of lymphoma cases: no other information | 214 | Healthy blood donors matched for age, gender, ethnicity and geographical location |
| Yri O et al, 2012 | Norway |
IL10 GSTP1 GSTT1 GSTM1 UGT1A1 |
Unspecified | 224 | HL patients: 135 males, 89 males: mean age 38 (18-84). | 1056 | Healthy blood donors (age and gender unknown) |
| Ruiz-Cosano J et al, 2012 | Spain |
GSTT1 GSTM1 |
Caucasian | 20 | HL a subset of lymphoma: no other information | 214 | Healthy blood donors matched for age, gender, ethnicity and geographical location |
| Monroy C et al, 2011 | USA | COX2 | Mixed | 200 | HL patients: mean age 47.2 years (SD 13.37 years). 107 males and 93 females. 80% NSHL. | 220 | Frequency matched to the age (within 5 years), sex, and race/ethnicity of the cases. Controls randomly selected random digit dialling. Mean age 49.27 years (SD 15.19 years). 123 males and 97 females. |
| Kasperzyk J et al, 2010 | USA |
MTHFR MTR |
Mixed (predominantly Caucasian) | 497 | HL patients: mean age 38 years (SD 15 years). EBV positive 16.3% and EBV negative 57.6%. NLPHL excluded from the study. | 638 | Frequency matched to the age (within 5 year age groups), sex, and state of residency distribution of the cases. Controls randomly selected through “town books” in Boston or random digit dialling in Connecticut. |
| Ribrag V et al, 2009 | France |
GSTT1 GSTM1 GSTP1 UGT1A1 |
Unspecified | 313 | HL patients: 180 males, 133 females. Median age 32 years (range 15-93 years). | 226 | Controls obtained from the French Blood Service. |
| Lourenco G et al, 2009 | Brazil |
GSTP1 GSTT1 GSTM1 |
Mixed | 110 | HL patients: median age 27 years (range 14-82 years), 57 males 53 females. Genotype data stratified by age, gender, ethnic origin and stage of disease. | 226 | Blood donors: median age 52 years (range 25-60 years), 151 males, 75 females. |
| El-Zein R et al, 2009 | USA | XPG/ERCC5 | Mixed | 200 | HL patients: mean age 47.2 years (SD 13.37 years). 107 males and 93 females. | 220 | Frequency matched to the age (within 5 years), sex, and race/ethnicity of the cases. Controls randomly selected random digit dialling. Mean age 49.27 years (SD 15.19 years). 123 males and 97 females. |
| Chang E et al, 2009 | USA | COX2 | Mixed | 473 | HL patients: 242 males, 231 females. NLPHL excluded from the study. | 373 | Frequency matched to the age (within 5 year age groups), sex, and state of residency distribution of the cases. Controls in Boston randomly selected through “town books”. Controls in Connecticut randomly selected through digit dialling (18-65 years of age) or Medicare files (66-79 years of age). 211 males and 162 females. |
| Hoeft K et al, 2008 | Germany | COX2 | Unspecified | 116 | HL a subset of lymphoma cases. | 710 | Randomly selected from the population registers of the study region. Matched for gender, age (1 year) and study region. 390 males and 320 females. |
| Timuragaoglu A et al, 2006 | Turkey | MTHFR | Unspecified | 30 | HL a subset of lymphoma cases. | 82 | Frequency matched to cases by age and gender. |
| Deligezer A et al, 2006 | Turkey | MTHFR | Unspecified | 51 | HL a subset of lymphoma cases. 22 cases MCHL, 20 NSHL, 9 LRHL. Median age 35 years (range 19-70), 35 males, 31 females. | 154 | Frequency matched to cases by age and gender of similar ethnic background. Randomly selected. |
| Nieters A et al, 2006 | Germany | IL10 | Unspecified | 115 | HL a subset of lymphoma cases. | 710 | 1:1 matched for gender, age (within 1 year) and study region. |
| Cordano et al, 1999 | UK | IL6 | Unspecified | 584 | HL patients: 61.3% NSHL and 21.4% cHL. 33% EBV positive. | 513 | Frequency matched to cases by age and gender, region of residence. |
| Cozen W et al, 2004 | USA | IL6 | Mixed (predominantly Caucasian) | 27 | HL patients: an affected twin of a monozygotic or dizygotic pair. Geneotype data stratified by zygosity and histology. | 201 | Spouses within 5 years of the twins’ ages, non-blood relative/friend or a age (within 5 years) or ethnicity-matched control subject was chosen from employees within the institution. |
| Munro L et al, 2003 | UK | IL10 | Unspecified | 125 | HL patients: mean age 44.04 years, 52.7% NSHL, 17.7 MCHL, 6.8% NLPHL, 2% LRCHL, 19. UC. 69 males, 88 females. Geneotype data stratified by histopathology. | 125 | Mean age 58.3 years, 51 males, 60 females. |
| Hohaus S et al, 2003 | Italy |
GSTT1 GSTM1 |
Unspecified | 90 | HL patients: median age 33 years, 54 males, 36 females. 67 NSHL, 7 MCHL, 6 NLPHL, 1 LDHL, 9 UC. Geneotype data stratified by age and gender. | 176 | Matched for sex and age (69 females, 107 males; median age 38 years, range 19–71 years) |
| Cunningham L et al, 2003 | Australia | IL10 | Mixed | 44 | HL a subset of lymphoma cases. | 164 | Geographically and ethnically similar metropolitan population. |
| Sarmanova J et al, 2001 | Norway |
GSTP1 GSTT1 GSTM1 |
Caucasian | 143 | HL a subset of lymphoma cases. Genotype data stratified by gender. | 455 | Similar gender and age distribution as overall lymphoma cohort. Staff from institute and nearby inhabitants of houses for elderly citizens. |
| Gonzalez Ordonez A et al, 2000 | Spain | MTHFR | Unspecified | 29 | HL a subset of lymphoma cases. Mean age 24 years (SD 15 years). | 200 | Healthy Spanish volunteers. |
| Lemos M et al, 1999 | Portugal | GSTM1 | Portuguese | 25 | HL a subset of lymphoma cases. | 128 | Unrelated Portuguese Caucasian volunteers; 56 males, 72 females; no history of cancer or other chronic disease; no age-matching. |
HL, Hodgkin Lymphoma; SD, standard deviation; EBV, Epstein-Barr Virus; cHL, Classical Hodgkin Lymphoma; NSHL, nodular sclerosis Hodgkin Lymphoma; MCHL, mixed cellularity Hodgkin Lymphoma; LRHL, lymphocyte-rich Hodgkin-Lymphoma; LDHL, lymphocyte-depleted Hodgkin-Lymphoma; NLPHL, nodular lymphocyte-predominant Hodgkin-Lymphoma; UC, unclassified
Table 3. Summary of odds ratios of individual studies along with their confidence intervals.
| Heterozygous model |
Homozygous model |
Carrier status |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 95% CIs |
95% CIs |
95% CIs |
|||||||||||
| Study | Polymorphism | OR | Upper | Lower | P | OR | Upper | Lower | P | OR | Upper | Lower | P |
| Yri | IL-10, rs1800890 | 0.95 | 1.30 | 0.69 | 0.74 | 1.41 | 2.17 | 0.94 | 0.12 | 1.04 | 1.40 | 0.77 | 0.80 |
| Nieters | IL-10, rs1800890 | 1.07 | 1.63 | 0.7 | 0.77 | 0.63 | 1.29 | 0.19 | 0.20 | 0.96 | 1.45 | 0.64 | 0.86 |
| Nieters | IL10, rs1800896 | 0.96 | 1.51 | 0.61 | 0.86 | 0.62 | 1.14 | 0.15 | 0.12 | 0.85 | 1.30 | 0.55 | 0.45 |
| Munro | IL10, rs1800896 | 0.84 | 1.48 | 0.47 | 0.54 | 0.83 | 1.68 | 0.44 | 0.61 | 0.84 | 1.43 | 0.49 | 0.51 |
| Cunningham | IL10, rs1800896 | 0.92 | 2.00 | 0.43 | 0.84 | 0.54 | 1.49 | 0.18 | 0.23 | 0.79 | 1.66 | 0.38 | 0.54 |
| Cordano | IL6, rs1800795 | 0.85 | 1.17 | 0.61 | 0.32 | 1.03 | 1.58 | 0.19 | 0.88 | 0.89 | 1.21 | 0.66 | 0.47 |
| Cozen | IL6, rs1800795 | 0.58 | 1.13 | 0.30 | 0.11 | 0.35 | 0.95 | 0.14 | 0.04 | 0.52 | 0.98 | 0.27 | 0.04 |
| Yri | GSTP1, rs1695 | 0.85 | 1.17 | 0.62 | 0.32 | 1.33 | 2.03 | 0.24 | 0.19 | 0.96 | 1.28 | 0.71 | 0.76 |
| Lourenco | GSTP1, rs1695 | 0.34 | 0.57 | 0.20 | 0.00 | 0.58 | 1.13 | 0.04 | 0.11 | 0.39 | 0.63 | 0.25 | 0.00 |
| Ribrag | GSTP1, rs1695 | 1.01 | 1.49 | 0.68 | 0.97 | 1.06 | 1.90 | 0.66 | 0.85 | 1.02 | 1.47 | 0.71 | 0.92 |
| Sarmanova | GSTP1, rs1695 | 0.77 | 1.33 | 0.44 | 0.35 | 1.88 | 3.70 | 0.38 | 0.06 | 1.00 | 1.61 | 0.61 | 0.98 |
| Yri | GSTT1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 3.17 | 5.09 | 1.97 | 0.00 |
| Ruiz-Cosano | GSTT1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 2.95 | 7.96 | 1.10 | 0.03 |
| Ribrag | GSTT1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 0.90 | 1.47 | 0.55 | 0.67 |
| Lourenco | GSTT1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 0.92 | 1.65 | 0.51 | 0.78 |
| Hohaus | GSTT1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 1.90 | 3.46 | 1.04 | 0.03 |
| Sarmanova | GSTT1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 0.88 | 1.79 | 0.43 | 0.72 |
| Yri | GSTM1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 1.18 | 1.57 | 0.88 | 0.27 |
| Ruiz-Cosano | GSTM1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 0.53 | 1.34 | 0.21 | 0.18 |
| Ribrag | GSTM1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 0.83 | 1.2 | 0.58 | 0.33 |
| Lourenco | GSTM1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 1.35 | 2.14 | 0.86 | 0.19 |
| Hohaus | GSTM1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 0.75 | 1.25 | 0.45 | 0.26 |
| Sarmanova | GSTM1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 0.90 | 1.45 | 0.55 | 0.66 |
| Lemos | GSTM1, deletion | NA | NA | NA | NA | NA | NA | NA | NA | 0.93 | 2.20 | 0.39 | 0.87 |
| Ruiz-Cosano | XPG/ERCC5, rs17655 | 1.89 | 5.28 | 0.68 | 0.22 | 4.86 | 18.69 | 3.81 | 0.01 | 2.33 | 6.06 | 0.89 | 0.08 |
| El-Zein | XPG/ERCC5, rs17655 | 1.19 | 1.79 | 0.79 | 0.40 | 1.63 | 3.60 | 0.58 | 0.22 | 1.25 | 1.84 | 0.85 | 0.26 |
| Kasperzyk | MTHFR, rs1801133 | 0.76 | 1.03 | 0.56 | 0.08 | 0.96 | 1.52 | 0.16 | 0.87 | 0.80 | 1.06 | 0.60 | 0.13 |
| Timuragaoglu | MTHFR, rs1801133 | 1.15 | 2.77 | 0.48 | 0.75 | 0.55 | 2.87 | 0.42 | 0.48 | 1.02 | 2.38 | 0.44 | 0.96 |
| Deligezer | MTHFR, rs1801133 | 0.49 | 0.96 | 0.25 | 0.04 | 0.26 | 1.19 | 0.07 | 0.06 | 0.45 | 0.85 | 0.23 | 0.01 |
| Gonzales Ordonez | MTHFR, rs1801133 | 0.77 | 1.76 | 0.33 | 0.53 | 0.92 | 3.48 | 0.34 | 0.9 | 0.8 | 1.73 | 0.36 | 0.56 |
| Ruiz-Cosano | MTR, rs1805087 | 0.84 | 2.70 | 0.26 | 0.78 | 5.81 | 21.90 | 2.06 | 0.00 | 1.47 | 3.73 | 0.58 | 0.41 |
| Kasperzyk | MTR, rs1805087 | 0.93 | 1.26 | 0.68 | 0.65 | 0.60 | 1.22 | 0.16 | 0.16 | 0.88 | 1.18 | 0.66 | 0.41 |
| Monroy | COX2, rs20417 | 1.39 | 0.74 | 2.6 | 0.31 | 6.94 | 2.68 | 59.16 | 0.04 | 1.6 | 2.93 | 0.87 | 0.13 |
| Chang | COX2, rs20417 | 1.11 | 1.49 | 0.82 | 0.51 | 1.99 | 4.24 | 0.65 | 0.07 | 1.18 | 1.57 | 0.88 | 0.27 |
| Hoeft | COX2, rs20417 | 1.05 | 1.65 | 0.67 | 0.83 | NA | NA | NA | NA | 0.99 | 1.55 | 0.63 | 0.97 |
| Chang | COX2, rs689466 | 0.99 | 1.36 | 0.73 | 0.97 | 1.16 | 2.39 | 0.33 | 0.69 | 1.01 | 1.36 | 0.75 | 0.93 |
| Hoeft | COX2, rs689466 | 0.99 | 1.53 | 0.63 | 0.95 | 0.31 | 2.35 | 0.31 | 0.23 | 0.93 | 1.43 | 0.60 | 0.73 |
| Yri | UGT1A1, tandem repeat | 0.90 | 1.22 | 0.66 | 0.49 | 0.79 | 1.32 | 0.16 | 0.37 | 0.88 | 1.17 | 0.66 | 0.37 |
| Ribrag | UGT1A1, tandem repeat | 0.70 | 1.04 | 0.47 | 0.07 | 0.71 | 1.32 | 0.12 | 0.28 | 0.70 | 1.02 | 0.48 | 0.06 |
CI, confidence interval; OR, odds ratio
Immune response genes
One of the two studies which examined the association between IL-10 rs1800890 and HL demonstrated a lower risk of HL for those >40 years with homozygosity or carrier status of the T allele (24). The departure of genotype frequencies from HWE (pHWE = 0.02) in the control group however suggests population stratification in this study. Pooling data from the both studies failed to demonstrate a significant association between the variant and HL risk (Table 4) (24, 28). In addition, there was no evidence of an association when pooling together three studies IL10, rs1800896 nor with the inclusion of data from the GWAS (Table 4) (7, 8, 24, 25, 29).
Table 4. Pooled odds ratios of all polymorphisms examined more than once in the literature including data from GWAS together with the false positive report probabilities (FPRP).
| Power | FPRP at prior probability of 0.001 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene, variant | Studies | Cases | Controls | OR | P | Phet | I2 | OR=1.2 | OR=1.5 | OR=1.2 | OR=1.5 |
| IL-10 rs1800890 | 2 | 334 | 1709 | ||||||||
| AT vs TT | 0.99 (0.77 - 1.28) | 0.93 | 0.67 | 0 | 0.91 | 1.00 | |||||
| AA vs TT | 0.99 (0.45 - 2.18) | 0.98 | 0.06 | 72% | 0.67 | 0.84 | |||||
| Carrier | 1.01 (0.79 - 1.29) | 0.93 | 0.77 | 0 | 0.92 | 1.00 | |||||
| IL10, rs1800896 | 5 | 1573 | 7026 | ||||||||
| AG vs AA | 1.01 (0.84 – 1.20) | 0.95 | 0.89 | 0 | 0.98 | 1.00 | |||||
| GG vs AA | 0.88 (0.50 – 1.54) | 0.65 | 0.01 | 73% | 0.58 | 0.84 | |||||
| Carrier | 1.06 (0.89 – 1.25) | 0.53 | 0.34 | 10% | 1.00 | 1.00 | |||||
| IL-6 rs1800795 | 2 | 435 | 374 | ||||||||
| GC vs GG | 0.79 (0.59 - 1.06) | 0.11 | 0.316 | 1% | 0.53 | 0.93 | |||||
| CC vs GG | 0.86 (0.59 - 1.29) | 0.50 | 0.05 | 74% | 0.78 | 0.97 | |||||
| Carrier | 0.92 (0.32 – 2.63) | 0.17 | <0.01 | 88.5% | 0.63 | 0.96 | |||||
| GSTP1, rs1695 | 6 | 2185 | 8287 | ||||||||
| AG vs GG | 0.82 (0.63 – 1.07) | 0.15 | <0.001 | 79% | 0.45 | 0.94 | |||||
| AA vs GG | 1.08 (0.92 – 1.27) | 0.35 | 0.06 | 52% | 0.90 | 1.00 | |||||
| Carrier | 0.88 (0.69 – 1.13) | 0.32 | <0.001 | 78% | 0.67 | 0.99 | |||||
| GSTT1 deletion | 6 | 796 | 2058 | ||||||||
| Carrier | 1.50 (0.90 - 2.50) | 0.12 | 0 | 76% | 0.20 | 0.50 | |||||
| GSTM1 deletion | 7 | 835 | 2205 | ||||||||
| Carrier | 0.99 (0.84-1.17) | 0.90 | 0.32 | 15% | 0.98 | 1 | |||||
| XPG/ERCC5, rs17655 | 3 | 1094 | 1651 | ||||||||
| GC vs GG | 0.94 (0.80 – 1.12) | 0.50 | 0.17 | 44% | 0.91 | 1.00 | |||||
| CC vs GG | 1.68 (1.18 – 2.37) | 0.01 | 0.27 | 23% | 0.03 | 0.26 | 0.99 | 0.99 | |||
| Carrier | 1.01 (0.86 – 1.19) | 0.88 | 0.10 | 57% | 0.65 | 0.82 | |||||
| MTHFR rs1801133 | 4 | 2025 | 7202 | ||||||||
| CT vs CC | 0.96 (0.85 – 1.07) | 0.47 | 0.19 | 33% | 0.99 | 1.00 | |||||
| TT vs CC | 0.98 (0.82 – 1.17) | 0.80 | 0.62 | 0 | 0.97 | 1.00 | |||||
| Carrier | 0.96 (0.86 – 1.07) | 0.45 | 0.15 | 38% | 1.00 | 1.00 | |||||
| MTR rs1805087 | 4 | 1936 | 6991 | ||||||||
| AG vs AA | 0.97 (0.86 – 1.10) | 0.63 | 0.99 | 0 | 0.99 | 1.00 | |||||
| GG vs AA | 1.18 (0.88 – 1.57) | 0.27 | 0.05 | 67% | 0.55 | 0.95 | |||||
| Carrier | 0.99 (0.88 – 1.11) | 0.86 | 0.72 | 0 | 0.99 | 1.00 | |||||
| COX2 rs20417 | 4 | 1538 | 2333 | ||||||||
| GC vs GG | 1.01 (0.92 – 1.24) | 0.41 | 0.83 | 0 | 0.95 | 1.00 | |||||
| CC vs GG | 1.37 (0.88 – 2.13) | 0.17 | 0.16 | 43% | 0.28 | 0.67 | |||||
| Carrier | 1.01 (0.94 – 1.26) | 0.27 | 0.50 | 0 | 0.94 | 1.00 | |||||
| COX2 rs689466 | 2 | 567 | 1024 | ||||||||
| AG vs AA | 0.991 (0.77 - 1.28) | 0.94 | 0.98 | 0 | 0.91 | 1 | |||||
| GG vs AA | 0.92 (0.48 - 1.76) | 0.81 | 0.22 | 33% | 0.62 | 0.84 | |||||
| Carrier | 0.98 (0.77 - 1.26) | 0.90 | 0.74 | 0 | 0.91 | 1 | |||||
| UGT1A1 TR | 2 | 532 | 1208 | ||||||||
| 6/7 vs 6/6 | 0.82 (0.64 - 1.040) | 0.10 | 0.33 | 0 | 0.44 | 0.95 | |||||
| 7/7 vs 6/6 | 0.76 (0.51 - 1.13) | 0.17 | 0.79 | 0 | 0.32 | 0.74 | |||||
| Carrier | 0.81 (0.64 - 1.01) | 0.06 | 0.36 | 0 | 0.38 | 0.95 | |||||
CI, confidence interval; OR, odds ratio
Two studies have evaluated rs1800795, a SNP located upstream of IL-6 (21, 23). One study utilised monozygotic and dizygotic twins with at least one affected twin. When all cases were analysed, homozygosity of the C allele appeared to be protective for HL and this was identified in the NS subgroup (21). These findings were not however, supported by a larger analysis of unrelated patients with HL nor in the pooled analysis (23) (Table 4).
Carcinogen metabolism genes
Four studies have investigated the SNP rs1696 in GSTP1 and risk of developing HL (18, 24, 30, 31). One study found a protective effect of the heterozygous (AG) genotype (24); while another study found the wild type genotype (AA) was associated with an increased risk of HL (18) and a third study found the less prevalent genotype (GG) to be associated with an increased risk of HL (30) (Table 3). Two studies demonstrated evidence of population stratification with departure from HWE in the controls (pHWE = 0.01). No evidence of association was found when these studies were pooled nor with the inclusion of data from the GWAS (Table 4).
Six studies have investigated deletion of the GSTT1 gene and predisposition to HL. GSTT1 deletion conferred an increased risk of HL in three studies (24, 27, 32) and no effect in three studies (18, 30, 31) (Table 3). Pooling data from the six studies revealed no evidence of association of GSTT1 deletion with HL risk (Table 4).
Seven studies have examined GSTM1 deletion and risk of developing HL (18, 24, 26, 30–33). Neither the studies nor a pooled analysis provides evidence for an association between GSTM1 deletion and HL risk (Table 3, Table 4).
DNA repair genes
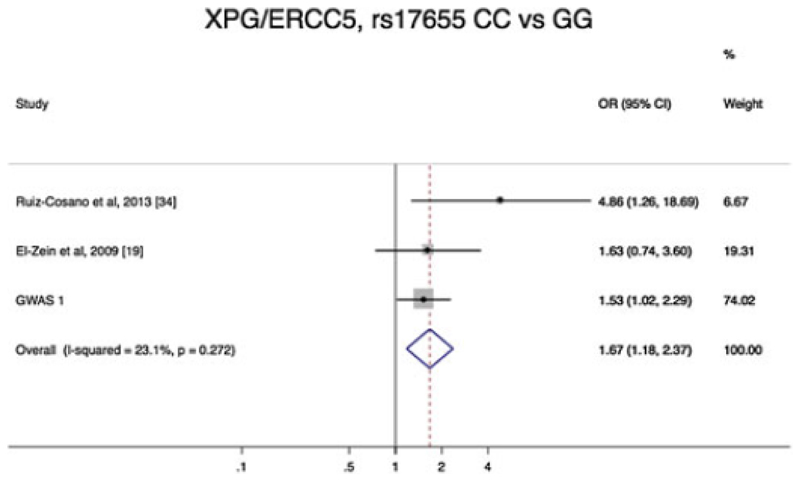
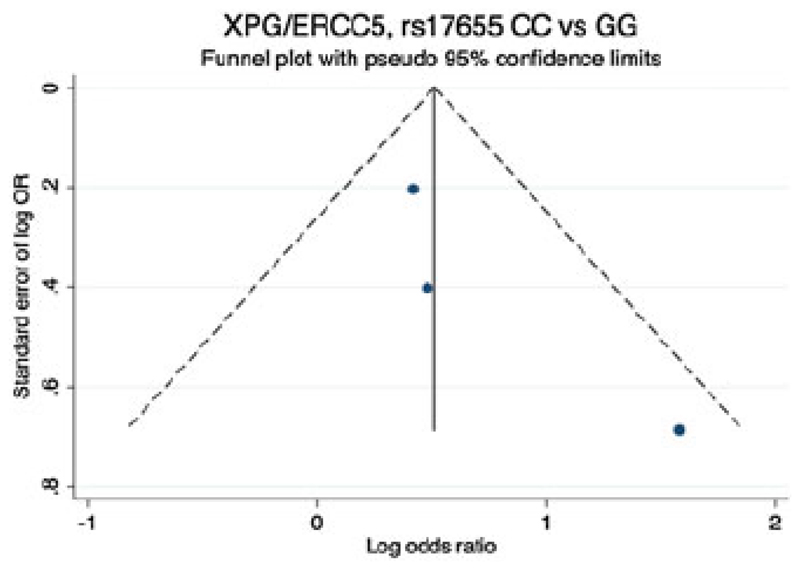
Two studies have examined the risk of HL and the SNP rs17655 in XPG/ERCC5 (19, 34), neither demonstrating an association between genotype and HL risk (Table 3). However, in a pooled analysis, a significant association between XPG/ERCC5, rs17655 CC homozygosity and an increased risk of developing HL with an OR=2.03 (95% CI:1.01 - 4.06, p=0.046) (Figure 2). Furthermore with the addition of data from GWAS the association appeared promising 1.68 (95% CI:1.18 – 2.37, p=0.01) (Figure 2 and Table 4). There was no evidence of small study effect (Figure 3).
Figure 2.
Forest plot of odds ratios (ORs) of Hodgkin lymphoma associated with polymorphic variant XPG/ERCC5 rs17655 CC versus GG. Boxes denote allelic OR point estimates, their areas being proportional to the inverse variance weight of the estimate. Horizontal lines represent 95% confidence intervals (CI). The diamond (and broken line) represents the summary OR computed under a fixed effects model, with 95% CI given by its width. The unbroken vertical line is at the null value (OR = 1.0)
Figure 3.
Begg’s funnel plot (using odds ratio (OR) of Hodgkin lymphoma risk associated with variant genotype rs17655 CC versus GG). The horizontal line represents the meta-analysis summary estimate, and the diagonal lines pseudo-95% CI limits about the effect estimate. In the absence of publication bias, studies will be distributed symmetrically above and below the horizontal line. Asymmetry on the top of the graph indicates evidence of publication bias towards studies reporting a positive logOR. LogOR, natural logarithm of the OR; s.e. of logOR, standard error of the logOR
Folate metabolism genes
Four studies have evaluated the risk of HL with rs1801133, in MTHFR (17, 35–37). In isolation, none of the studies provided evidence for an association between the genotype and risk (Table 3). However, in a pooled analysis, heterozygosity conferred an OR of 0.74 (95% CI:0.578 - 0.95, p=0.018) and carrier status an OR of 0.75 (95% CI: 0.60 - 0.96, p=0.019). However, with the addition of GWAS data no association was demonstrated (Table 4).
Two studies have evaluated the risk of HL with rs1805087 in MTR (17, 34). Neither demonstrated an association (Table 3). Similarly no association was shown in a meta-analysis which included GWAS data (Table 4).
Other Genes
Three studies have investigated the role of rs20417 in COX2 and HL risk (20, 22, 38). One study demonstrated an association with both carrier and homozygous minor genotype (20) (Table 3). However in a pooled analysis with the inclusion of data from GWAS no association was seen (Table 4). Two studies have also examined the risk of HL rs689466 in COX2, but no association is seen (Table 4) (22, 38).
Finally, two studies have investigated the tandem repeat in UGT1A1 and HL risk. No significant association was seen in either study (Table 3). A pooled analysis did not provide evidence for a relationship between UGT1A1 genotype and HL (Table 4) (24, 31).
Variants examined once in published studies
Using directly typed SNPs from the GWAS, we were able to include an additional 47 variants published in eight papers (39–46). There was evidence for association in eight of these variants (p<0.05).
False Positive Report Probability
To evaluate the robustness of the three significant findings from the pooled analyses, we calculated FPRP conditional on a prior probability of 0.001. None of the above results are not considered noteworthy on the basis of the pre-defined assumptions (Table 4 and Table 5). For example, although the summary OR from the pooled analysis of rs17655 indicated a statistically significant positive association with risk, the FPRP was 0.99 which is much higher than the conventionally accepted threshold cut off for noteworthiness of <0.2.
Table 5. Pooled odds ratios of all the polymorphisms examined once in the literature and GWAS data with the false positive report probabilities (FPRP).
| Power | FPRP at prior probability of 0.001 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene, variant | Studies | Cases | Controls | OR | P | Phet | I2 | OR=1.2 | OR=1.5 | OR=1.2 | OR=1.5 |
| TNFα, rs1800750 | 2 | 928 | 1345 | ||||||||
| GA vs GG | 1.78 (0.34 – 9.19) | 0.50 | 0.01 | 84% | 0.32 | 0.42 | |||||
| AA vs GG | |||||||||||
| Carrier | 1.78 (0.34 – 9.19) | 0.50 | 0.01 | 84% | 0.32 | 0.42 | |||||
| TNFα, rs1800629 | 2 | 987 | 1893 | ||||||||
| GA vs GG | 1.14 (0.94 – 1.37) | 0.19 | 0.66 | 0 | 0.71 | 1.00 | |||||
| AA vs GG | 1.17 (0.74 – 1.87) | 0.50 | 0.43 | 0 | 0.54 | 0.85 | |||||
| Carrier | 1.14 (0.95 – 1.37) | 0.15 | 0.81 | 0 | 0.71 | 1.00 | |||||
| FCGR2, rs1801274 | 3 | 1704 | 7938 | ||||||||
| CT vs CC | 1.15 (1.00 - 1.31) | 0.05 | 0.93 | 0 | 0.74 | 1.00 | |||||
| TT vs CC | 1.07 (0.91 - 1.26) | 0.40 | 0.37 | 0 | 0.92 | 1.00 | |||||
| Carrier | 1.12 (0.99 - 1.27) | 0.08 | 0.96 | 0 | 0.86 | 1.00 | |||||
| IL1A, rs1800587 | 2 | 977 | 1877 | ||||||||
| CT vs CC | 0.85 (0.72 – 1.00) | 0.05 | 0.87 | 0 | 0.59 | 1.00 | |||||
| TT vs CC | 0.72 (0.54 – 0.96) | 0.02 | 0.10 | 63% | 0.70 | 0.99 | 1.00 | 1.00 | |||
| Carrier | 1.00 (0.60 – 1.67) | 0.99 | 0.03 | 80% | 0.94 | 1.00 | |||||
| IL1B, rs16944 | 2 | 981 | 1876 | ||||||||
| GA cs GG | 1.03 (0.87 – 1.22) | 0.73 | 0.17 | 47% | 0.96 | 1.00 | |||||
| AA vs GG | 1.36 (1.05 – 1.77) | 0.02 | 0.329 | 9% | 0.18 | 0.77 | 0.99 | 0.97 | |||
| Carrier | 1.09 (0.93 – 1.28) | 0.30 | 0.41 | 0 | 0.88 | 1.00 | |||||
| IL-4 rs2243248 | 3 | 1685 | 7465 | ||||||||
| TG vs GG | 0.97 (0.70 - 1.33) | 0.83 | 0.04 | 70% | 0.83 | 0.99 | |||||
| GG vs TT | 0.88 (0.42 - 1.86) | 0.50 | 0.44 | 0 | 1.00 | 1.00 | |||||
| Carrier | 1.02 (0.86 – 1.20) | 0.84 | 0.08 | 61% | 0.98 | 1.00 | |||||
| IL-4 rs2243250 | 2 | 976 | 1318 | ||||||||
| CT vs CC | 1.06 (0.87 - 1.28) | 0.58 | 0.91 | 0 | 0.90 | 1.00 | |||||
| TT vs CC | 1.17 (0.69 – 2.00) | 0.57 | 0.08 | 68% | 0.54 | 0.82 | |||||
| Carrier | 1.29 (0.77 – 2.16) | 0.33 | 0.05 | 75% | 0.39 | 0.72 | |||||
| IL4A rs1801275 | 3 | 1565 | 6517 | ||||||||
| AG vs AA | 0.87 (0.76 – 0.99) | 0.03 | 0.0 | 0 | 0.74 | 1.00 | 0.98 | 0.97 | |||
| GG vs AA | 1.07 (0.80 -1.43) | 0.65 | 0.29 | 18% | 0.78 | 0.99 | |||||
| carrier | 0.89 (0.79 – 1.01) | 0.07 | 0.70 | 0 | 0.85 | 1.00 | |||||
| IL10, rs1800872 | 2 | 987 | 1893 | ||||||||
| CA vs CC | 0.98 (0.82 – 1.16) | 0.79 | 0.09 | 64% | 0.97 | 1.00 | |||||
| AA vs CC | 1.26 (0.82 – 1.93) | 0.30 | 0.65 | 0 | 0.41 | 0.79 | |||||
| Carrier | 1.00 (0.85 – 1.20) | 0.96 | 0.16 | 49% | 0.98 | 1..00 | |||||
| IL10RA, rs2229113 | 3 | 986 | 1890 | ||||||||
| CT vs CC | 0.85 (0.72 – 1.01) | 0.06 | 0.97 | 0 | 0.59 | 1.00 | |||||
| TT vs CC | 0.81 (0.62 – 1.06) | 0.12 | 0.88 | 0 | 0.40 | 0.96 | |||||
| Carrier | 0.84 (0.72 – 0.99) | 0.04 | 0.93 | 0 | 0.54 | 1.00 | 0.97 | 0.97 | |||
| IL10RA, rs3135932 | 3 | 987 | 1893 | ||||||||
| AG vs AA | 0.98 (0.82 – 1.16) | 0.76 | 0.09 | 64% | 0.97 | 1.00 | |||||
| GG vs AA | 1.26 (0.82 – 1.93) | 0.30 | 0.65 | 0 | 0.41 | 0.79 | |||||
| Carrier | 1.26 (0.46 – 3.48) | 0.65 | <0.001 | 94% | 0.46 | 0.63 | |||||
| IRF4, rs872071 | 3 | 1592 | 7442 | ||||||||
| AG vs GG | 0.92 (0.81 – 1.04) | 0.19 | 0.41 | 0 | 0.94 | 1.00 | |||||
| AA vs GG | 0.88 (0.76 – 1.02) | 0.09 | 0.39 | 0 | 0.77 | 1.00 | |||||
| Carrier | 0.90 (0.80 – 1.02) | 0.09 | 0.52 | 0 | 0.89 | 1.00 | |||||
| LTC4S, rs730012 | 3 | 1915 | 6778 | ||||||||
| CA vs AA | 0.94 (0.84 – 1.06) | 0.34 | 0.41 | 0 | 0.98 | 1.00 | |||||
| CC vs AA | 0.97 (0.79 – 1.19) | 0.75 | 0.44 | 0 | 0.93 | 1.00 | |||||
| CArrier | 0.95 (0.85 – 1.06) | 0.35 | 0.44 | 0 | 0.99 | 1.00 | |||||
| CXCL12, rs1801157 | 2 | 912 | 1308 | ||||||||
| GA vs GG | 1.05 (0.87 – 1.23) | 0.60 | 0.97 | 0 | 0.95 | 1.00 | |||||
| AA vs GG | 1.26 (0.78 – 2.02) | 0.34 | 0.43 | 0 | 0.42 | 0.77 | |||||
| Carrier | 1.07 (0.89 – 1.28) | 0.46 | 0.75 | 0 | 0.90 | 1.00 | |||||
| TLR1, rs5743551 | 2 | 985 | 1881 | ||||||||
| AG vs AA | 0.90 (0.76 – 1.06) | 0.21 | 0.12 | 59% | 0.82 | 1.00 | |||||
| GG vs AA | 1.02 (0.72 – 1.45) | 0.91 | 0.95 | 0 | 0.82 | 0.98 | |||||
| Carrier | 0.91 (0.77 – 1.07) | 0.27 | 0.16 | 50% | 0.57 | 0.74 | |||||
| TLR2, rs4696480 | 2 | 986 | 1982 | ||||||||
| TA vs TT | 1.47 (0.89 – 2.45) | 0.13 | 0.05 | 75% | 0.22 | 0.53 | |||||
| AA vs TT | 1.09 (0.86 – 1.37) | 0.48 | 0.20 | 40% | 0.80 | 1.00 | |||||
| Carrier | 1.42 (0.85 – 2.37) | 0.18 | 0.04 | 76% | 0.26 | 0.59 | |||||
| TLR9, rs187084 | 2 | 986 | 1982 | ||||||||
| TC vs TT | 0.97 (0.81 – 1.16) | 0.71 | 0.63 | 0 | 0.95 | 1.00 | |||||
| CC vs TT | 0.83 (0.65 – 1.04) | 0.11 | 0.99 | 0 | 0.49 | 0.97 | |||||
| Carrier | 0.93 (0.78 – 1.10) | 0.72 | 0.34 | 0 | 0.90 | 1.00 | |||||
| TLR9, rs5743836 | 2 | 986 | 1982 | ||||||||
| TC vs TT | 1.11 (0.91 – 1.34) | 0.31 | 0.08 | 68% | 0.79 | 1.00 | |||||
| CC vs TT | 1.02 (0.80 – 1.30) | 0.89 | 0.99 | 0% | 0.91 | 0.99 | |||||
| Carrier | 1.07 (0.89 – 1.23) | 0.50 | 0.12 | 59% | 0.95 | 1.00 | |||||
| GSTA1, rs3957357 | 3 | 1688 | 7403 | ||||||||
| CT vs CC | 1.07 (0.91 – 1.25) | 0.41 | 0.19 | 40% | 0.93 | 1.00 | |||||
| TT vsCC | 0.81 (0.55 – 1.19) | 0.28 | 0.67 | 0 | 0.44 | 0.84 | |||||
| Carrier | 1.04 (0.89 – 1.21) | 0.62 | 0.23 | 33% | 0.97 | 1.00 | |||||
| EPHX, rs1051740 | 2 | 953 | 1665 | ||||||||
| TC vs TT | 0.98 (0.83 – 1.17) | 0.83 | 0.28 | 14% | 0.96 | 1.00 | |||||
| CC vs TT | 2.76 (0.20 – 38.40) | 0.45 | <0.001 | 97% | 0.27 | 0.33 | |||||
| Carrier | 1.14 (0.97 – 1.35) | 0.11 | 0.06 | 72% | 0.72 | 1.00 | |||||
| EPHX, rs2234922 | 2 | 666 | 5652 | ||||||||
| AG vs AA | 0.87 (0.73 – 1.03) | 0.11 | 0.78 | 0 | 0.70 | 1.00 | |||||
| GG vs AA | 0.93 (0.61 – 1.42) | 0.73 | 0.34 | 0 | 0.70 | 0.94 | |||||
| Carrier | 0.87 (0.73 – 1.03) | 0.11 | 0.58 | 0 | |||||||
| CYP2C9, rs1057910 | 2 | 1054 | 5570 | ||||||||
| AC vs AA | 1.26 (1.02 – 1.54) | 0.03 | 0.97 | 0 | 0.70 | 1.00 | 0.99 | 0.96 | |||
| CC vs AA | 1.33 (0.49 – 3.67) | 0.58 | 0.31 | 4% | 0.42 | 0.59 | |||||
| Carrier | 1.30 (1.03 – 1.54) | 0.03 | 0.87 | 0 | 0.18 | 0.95 | 0.93 | 0.72 | |||
| CYP2C9, rs1799853 | 2 | 1059 | 5569 | ||||||||
| CT vs CC | 1.28 (1.07 – 1.51) | 0.01 | 0.21 | 38% | 0.22 | 0.97 | 0.94 | 0.78 | |||
| TT vs CC | 0.34 (0.02 – 6.89) | 0.48 | 0.04 | 75% | 0.28 | 0.33 | |||||
| Carrier | 1.24 (1.05 – 1.46) | 0.01 | 0.41 | 0 | 0.35 | 0.99 | 1.00 | 0.99 | |||
| XPA, rs1800975 | 2 | 896 | 1432 | ||||||||
| GA vs GG | 0.92 (0.76 – 1.10) | 0.37 | 0.13 | 57% | 0.86 | 1.00 | |||||
| AA vs GG | 3.60 (0.49 – 26.70) | 0.21 | 0.01 | 86% | 0.14 | 0.20 | |||||
| Carrier | 1.02 (0.86 – 1.22) | 0.25 | 0.06 | 72% | 0.96 | 1.00 | |||||
| XPC, rs2228000 | 2 | 984 | 1878 | ||||||||
| GA vs GG | 0.84 (0.71 – 1.00) | 0.05 | 0.53 | 0 | 0.52 | 0.89 | |||||
| AA vs GG | 0.89 (0.70 – 1.50) | 0.47 | 0.14 | 46% | 0.60 | 0.86 | |||||
| Carrier | 0.85 (0.71 – 1.00) | 0.05 | 0.99 | 0 | 0.59 | 1.00 | |||||
| XPC, rs2228001 | 2 | 1663 | 6637 | ||||||||
| GT vs TT | 0.98 (0.86 – 1.12) | 0.78 | 0.97 | 0 | 0.99 | 1.2 | |||||
| GG vs TT | 1.01 (0.85 – 1.20) | 0.93 | 0.33 | 10% | 0.98 | 1.00 | |||||
| Carrier | 0.99 (0.86 – 1.12) | 0.84 | 0.88 | 0 | 0.91 | 1.00 | |||||
| ERCC1, rs3212986 | 3 | 1633 | 6715 | ||||||||
| CA vs CC | 0.80 (0.69 – 0.91) | 0.001 | 0.85 | 0 | 0.27 | 1.00 | 0.72 | 0.41 | |||
| AA vs CC | 0.87 (0.68 – 1.22) | 0.28 | 0.22 | 35% | 0.60 | 0.94 | |||||
| Carrier | 0.81 (0.72 – 0.91) | 0.001 | 0.98 | 0 | 0.32 | 1.00 | 0.56 | 0.28 | |||
| XRCC1, rs1799782 | 2 | 1072 | 1436 | ||||||||
| CT vs CC | 0.69 (0.28 – 1.71) | 0.42 | 0.01 | 84% | 0.35 | 0.53 | |||||
| TT vs CC | 0.73 (0.26 – 2.10) | 0.56 | 0.38 | 0 | 0.40 | 0.57 | |||||
| Carrier | 0.66 (0.25 – 1.74) | 0.42 | 0.01 | 86% | 0.32 | 0.50 | |||||
| XRCC3, rs861539 | 3 | 912 | 1308 | ||||||||
| GA vs GG | 0.97 (0.86 – 1.10) | 0.68 | 0.30 | 17% | 0.99 | 1.00 | |||||
| AA vs GG | 1.05 (0.88 – 1.26) | 0.57 | 0.74 | 0 | 0.92 | 1.00 | |||||
| Carrier | 0.99 (0.88 – 1.12) | 0.89 | 0.34 | 7% | 1.00 | 1.00 | |||||
| COX2 rs5277 | 2 | 1334 | 1577 | ||||||||
| CG vs CC | 1.07 (0.90 – 1.27) | 0.45 | 0.36 | 0 | 0.91 | 1.00 | |||||
| GG vs CC | 0.79 (0.50 – 1.25) | 0.31 | 0.73 | 0 | 0.41 | 0.77 | |||||
| Carrier | 1.01 (0.86 – 1.19) | 0.92 | 0.69 | 0 | 0.98 | 1.00 | |||||
| ABCC2, rs17222723 | 3 | 1688 | 6517 | ||||||||
| AT vs TT | 0.97 (0.79 – 1.20) | 0.80 | 0.74 | 0 | 0.92 | 1.00 | |||||
| AA vs TT | 2.02 (0.85 – 4.77) | 0.11 | 0.19 | 40% | 0.12 | 0.25 | |||||
| Carrier | 1.01 (0.83 – 1.23) | 0.94 | 0.95 | 0 | 0.96 | 1.00 | |||||
| NBN, rs1801282 | 2 | 1044 | 1435 | ||||||||
| CA vs CC | 1.07 (0.89 – 1.29) | 0.45 | 0.06 | 70% | 0.95 | 1.00 | |||||
| AA cs CC | 0.99 (0.61 – 1.60) | 0.95 | 0.21 | 37% | 0.76 | 0.75 | |||||
| Carrier | 1.07 (0.89 – 1.27) | 0.50 | 0.19 | 42% | 0.71 | 0.95 | |||||
| TP53, rs104522 | 2 | 1174 | 1967 | ||||||||
| GC vs CC | 1.12 (0.96 – 1.30) | 0.16 | 0.65 | 0 | 0.81 | 1.00 | |||||
| GG vs CC | 0.96 (0.71 – 1.29) | 0.76 | 0.54 | 0 | 0.83 | 0.99 | |||||
| Carrier | 1.09 (0.94 – 1.26) | 0.25 | 0.57 | 0 | |||||||
| NFKB1, rs3774937 | 2 | 1041 | 5565 | ||||||||
| TC vs TT | 1.02 (0.86 – 1.18) | 0.92 | 0.65 | 0 | 0.90 | 1.00 | |||||
| CC vs TT | 1.13 (0.90 – 1.44) | 0.29 | 0.46 | 0 | 0.69 | 0.99 | |||||
| Carrier | 0.85 (0.56 – 1.30) | 0.45 | 0.01 | 86% | 0.54 | 0.87 | |||||
| NFKB1A, rs696 | 2 | 1052 | 5563 | ||||||||
| AG vs GG | 1.24 (1.06 – 1.45) | 0.01 | 0.56 | 0 | 0.34 | 0.99 | 0.95 | 0.88 | |||
| AA vs GG | 1.40 (1.12 – 1.74) | 0.003 | 0.25 | 23% | 0.08 | 0.733 | 0.97 | 0.77 | |||
| Carrier | 1.27 (1.10 – 1.48) | 0.002 | 0.35 | 0 | 0.23 | 0.98 | 0.90 | 0.70 | |||
| NFKB1A, rs8904 | 2 | 1046 | 5566 | ||||||||
| CT cs CC | 1.13 (0.96 – 1.32) | 0.14 | 0.20 | 40% | 0.78 | 1.00 | |||||
| TT vs CC | 1.11 (0.89 – 1.39) | 0.36 | 0.05 | 75% | 0.77 | 1.00 | |||||
| Carrier | 1.12 (0.97 – 1.31) | 0.13 | 0.08 | 67% | 0.81 | 1.00 | |||||
| NFKB1A, rs1050851 | 2 | 1331 | 1579 | ||||||||
| CT vs CC | 0.87 (0.50 – 1.48) | 0.60 | 0.002 | 90% | 0.56 | 0.84 | |||||
| TT vs CC | 0.88 (0.61 – 1.28) | 0.51 | 0.52 | 0 | 0.61 | 0.93 | |||||
| Carrier | 0.87 (0.52 – 1.44) | 0.58 | 0.002 | 89% | 0.57 | 0.85 | |||||
| NFKB1A, rs19571006 | |||||||||||
| AT vs AA | 1.09 (0.93 – 1.27) | 0.30 | 0.90 | 0 | 0.89 | 1.00 | |||||
| TT vs AA | 1.29 (0.67 – 2.48) | 0.45 | 0.04 | 75% | 0.41 | 0.67 | |||||
| Carrier | 1.09 (0.94 – 1.27) | 0.24 | 0.61 | 0 | 0.89 | 1.00 | |||||
| CHUK, rs2230804 | 2 | 1339 | 1577 | ||||||||
| AG vs AA | 0.81 (0.54 – 1.24) | 0.33 | 0.03 | 78% | 0.45 | 0.82 | |||||
| GG vs AA | 0.75 (0.48 – 1.17) | 0.21 | 0.04 | 75% | 0.32 | 0.70 | |||||
| Carrier | 0.80 (0.52 – 1.22) | 0.29 | 0.02 | 82% | 0.43 | 0.80 | |||||
| PTGES, rs10448290 | 2 | 986 | 1884 | ||||||||
| AC vs AA | 0.99 (0.80 – 1.23) | 0.93 | 0.79 | 0 | 0.94 | 1.00 | |||||
| CC vs AA | 1.42 (0.61 – 3.31) | 0.41 | 0.06 | 71% | 0.35 | 0.55 | |||||
| Carrier | 1.01 (0.81 – 1.24) | 0.96 | 0.48 | 0 | 0.95 | 1.00 | |||||
| PTGES, rs2241270 | 2 | 700 | 5866 | ||||||||
| AC vs AA | 0.99 (0.82 – 1.20) | 0.93 | 0.84 | 0 | 0.97 | 1.00 | |||||
| CC vs AA | 0.90 (0.48 – 1.68) | 0.73 | 0.77 | 0 | 0.60 | 0.83 | |||||
| Carrier | 0.94 (0.82 – 1.18) | 0.87 | 0.79 | 0 | 0.85 | 1.00 | |||||
| PTGES, rs4837404 | 2 | 1326 | 1579 | ||||||||
| AG vs AA | 1.10 (0.94 – 1.29) | 0.25 | 0.87 | 0 | 0.78 | 0.99 | |||||
| GG vs AA | 1.08 (0.85 – 1.34) | 0.51 | 0.41 | 0 | 0.83 | 1.00 | |||||
| Carrier | 1.09 (0.94 – 1.27) | 0.24 | 0.92 | 0 | 0.89 | 1.00 | |||||
| HPSE, rs4693608 | 3 | 1481 | 6520 | ||||||||
| GA vs GG | 0.98 (0.84 – 1.14) | 0.77 | 0.91 | 0 | 0.98 | 1.00 | |||||
| AA vs GG | 0.98 (0.82 – 1.15) | 0.70 | 0.88 | 0 | 0.98 | 1.00 | |||||
| Carrier | 0.98 (0.85 – 1.13) | 0.76 | 0.86 | 0 | 0.99 | 1.00 | |||||
| HPSE, rs11099592 | 3 | 1483 | 6520 | ||||||||
| GA vs GG | 1.09 (0.96 – 1.24) | 0.17 | 0.51 | 0 | 0.93 | 1.00 | |||||
| AA vs GG | 0.87 (0.66 – 1.16) | 0.35 | 0.30 | 16% | 0.62 | 0.97 | |||||
| Carrier | 1.06 (0.94 – 1.20) | 0.34 | 0.39 | 0 | 0.98 | 1.00 | |||||
| HPSE, rs436425 | 2 | 607 | 5302 | ||||||||
| TC vs TT | 1.00 (0.82 – 1.20) | 0.97 | 0.60 | 0 | 0.98 | 1.00 | |||||
| CC vs TT | 0.90 (0.66 – 1.23) | 0.51 | 0.26 | 21% | 0.69 | 0.97 | |||||
| Carrier | 0.98 (0.83 – 1.16) | 0.81 | 0.94 | 0 | 0.97 | 1.00 | |||||
| UGT1A6, rs1105879 | 3 | 1929 | 6774 | ||||||||
| TC vs CC | 0.96 (0.85 – 1.08) | 0.46 | 0.40 | 0 | 0.99 | 1.00 | |||||
| CC vs TT | 1.02 (0.85 – 1.21) | 0.87 | 0.74 | 0 | 0.97 | 1.00 | |||||
| Carrier | 0.97 (0.87 – 1.09) | 0.58 | 0.60 | 0 | 1.00 | 1.00 | |||||
| UGT1A6, rs2070959 | 3 | 1923 | 6785 | ||||||||
| AG vs AA | 0.97 (0.86 – 1.09) | 0.61 | 0.54 | 0 | 1.00 | 1.00 | |||||
| GG vs AA | 1.07 (0.89 – 1.28) | 0.49 | 0.66 | 0 | 0.86 | 1.00 | |||||
| Carrier | 1.77 (0.56 – 5.52) | 0.33 | <0.001 | 99% | 0.25 | 0.39 | |||||
CI, confidence interval; OR, odds ratio
Discussion
It is clear that substantial research has been carried out examining polymorphic variants in a number of putative candidate genes as risk factors for HL. While our meta-analysis provides some support for a variation in XPG/ERCC5 as a risk factors for HL, as well as 8 other variants, these data should be interpreted with caution as the identified associations are not robust on the basis of multiple testing correction and FPRP.
Even excluding this, a number of general conclusions can be constructed from the published studies. Few of the studies variants have been reported as statistically significant in more than one study. It is generally acknowledged that independent replication of study findings is a prerequisite to assess the robustness of findings. In some studies, the failure to demonstrate a relationship may simply be a consequence of poor power because of sample size. Genome-wide association studies (GWAS) of cancer have revealed that the relative risk associated with common variants is, typically between 1.1 and 1.3(47). Fewer than 40% of the studies we reviewed had 80% power to demonstrate even a 2-fold difference in risk at the 0.05 significance level. To overcome this lack of power, we have undertaken a meta-analysis pooling the data from the published studies. There are, however, caveats to this statistical procedure.
In any systematic review, publication bias is clearly of great concern. The most common scenario is that negative findings may go unreported. Furthermore, many studies excluded do not describe the ethnicity of cases or controls, and it is assumed that each polymorphism is functional with respect to risk in each study population. If, however, the polymorphism is a neutral marker for another variant, the assumption may well not apply, since LD is often population-dependent. Here we have relied on data extractable from published reports. Ideally, access to primary data is desirable; in the absence of this it would be advisable that in the future at least summary data be published to allow meta-analysis to be conducted.
An important lesson from the published studies is that greater attention should be paid to study design. Data from GWAS have demonstrated a differing allelic architecture of genetic susceptibility to HL with respect to histology and EBV status. Few studies have stratified genotype data according to histology and EBV status. This may explain the lack of consistency in candidate gene association studies and in our meta-analysis. Due to the lack of stratification in the majority of studies we were unable to include this in our meta-analysis. The issue of population stratification in case-control studies and resulting false positive results is also of concern. Such associations occur because of population subdivision and non-random mating, leading to variation in the marker frequency within the population as a result of founder effects and/or genetic drift. The severity of spurious association becomes an increasing problem with increasing study size. To avoid this problem, potential confounding effect of population stratification should be allowed for in the design and analysis of the study. This requires the identification of sub-populations in terms of factors that can influence both disease and marker allele frequencies. Provided cases and controls are well matched, differences in the frequency of genotypes will only be seen at predisposition loci. Hence, stratification can be detected by typing a series of unlinked markers chosen from a panel known to exhibit differences in allele frequency between populations.
We have attempted to review published analyses of the relationship between polymorphic variation and risk of HL through several iterations of search criteria; it is possible, however, that we have missed some published studies. As the number of articles on genetic variation on risk of HL published in the past decade has increased considerably and continues to grow, we accept that this review provides a snapshot of progress to date in the field.
All of the studies we have reviewed have been based on a candidate gene approach. It is clear from studies of cancer that without a clear understanding of tumour causalities the definition of what constitutes a candidate gene is inherently problematic, making an unbiased approach through GWAS highly desirable. Moreover, the possibility of missing the identification of important variants in hitherto unstudied genes is avoided. Thus far GWAS of HL have provided evidence that variation in a number of genes including REL, EOMES, ERAP1, IL13, PVT1, GATA3 and TCF3 (7, 8, 48, 49) influence the risk of developing HL. In contrast to the candidate gene studies, the substantial evidence supporting these variants, including sizeable power and replication in large samples, indicate that the associations are highly robust. These data thus provide the first unambiguous evidence that common low penetrance susceptibility alleles contribute to the risk of HL.
Conclusions
The search for polymorphic variants influencing the risk of HL is a worthy enterprise. However, the studies that have been conducted to date have important lessons for the design and execution of future studies. Candidate gene analyses should be viewed as complementary to GWAS, as they theoretically offer advantages both in terms of statistical power and an ability to identify low frequency risk variants. Furthermore, many functional variants, such as the small scale insertion and deletions in carcinogen metabolism genes, are poorly captured by tagging SNPs used in GWAS. It is however, clear that in addition to conducting studies using adequate sample, attention should be paid to study design to avoid problems of aetiological heterogeneity, population stratification and other sources of potential bias in order to maximise the output of any future candidate gene study.
Acknowledgements
AS is in receipt of a Cancer Research UK Clinical Training Fellowship through The Institute of Cancer Research.
Footnotes
Ethical consideration: No experiments on human subjects were carried out as part of this research article. This article conforms with the host Institution ethical guidelines and the Declaration of Helsinki
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Howlander N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, et al. SEER Cancer Statistics Review, 1975-2011. Bethesda, MD: National Cancer Institute; 2013. [Google Scholar]
- 2.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri ES, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissue. 4th ed. Lyon, France: IARC Press; 2008. [Google Scholar]
- 3.Mack TM, Cozen W, Shibata DK, Weiss LM, Nathwani BN, Hernandez AM, et al. Concordance for Hodgkin's disease in identical twins suggesting genetic susceptibility to the young-adult form of the disease. The New England journal of medicine. 1995;332(7):413–8. doi: 10.1056/NEJM199502163320701. [DOI] [PubMed] [Google Scholar]
- 4.Shugart YY, Hemminki K, Vaittinen P, Kingman A, Dong C. A genetic study of Hodgkin's lymphoma: an estimate of heritability and anticipation based on the familial cancer database in Sweden. Human genetics. 2000;106(5):553–6. doi: 10.1007/s004390000291. [DOI] [PubMed] [Google Scholar]
- 5.Moutsianas L, Enciso-Mora V, Ma YP, Leslie S, Dilthey A, Broderick P, et al. Multiple Hodgkin lymphoma-associated loci within the HLA region at chromosome 6p21.3. Blood. 2011;118(3):670–4. doi: 10.1182/blood-2011-03-339630. [DOI] [PubMed] [Google Scholar]
- 6.Higgins J, Green Se. In: Cochrane Handbook for Systematic Reviews of Interventions Version 5.0.2. H JPT, G S, editors. The Cochrane Collaboration; 2009. [updated September 2009] [Google Scholar]
- 7.Frampton M, da Silva Filho MI, Broderick P, Thomsen H, Forsti A, Vijayakrishnan J, et al. Variation at 3p24.1 and 6q23.3 influences the risk of Hodgkin's lymphoma. Nature communications. 2013;4 doi: 10.1038/ncomms3549. 2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enciso-Mora V, Broderick P, Ma Y, Jarrett RF, Hjalgrim H, Hemminki K, et al. A genome-wide association study of Hodgkin's lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3) Nature genetics. 2010;42(12):1126–30. doi: 10.1038/ng.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bin Q, Luo J. Role of polymorphisms of GSTM1, GSTT1 and GSTP1 Ile105Val in Hodgkin and non-Hodgkin lymphoma risk: a Human Genome Epidemiology (HuGE) review. Leukemia & lymphoma. 2013;54(1):14–20. doi: 10.3109/10428194.2012.706284. [DOI] [PubMed] [Google Scholar]
- 10.Skibola CF, Curry JD, Nieters A. Genetic susceptibility to lymphoma. Haematologica. 2007;92(7):960–9. doi: 10.3324/haematol.11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute. 1959;22(4):719–48. [PubMed] [Google Scholar]
- 12.DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled clinical trials. 1986;7(3):177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 13.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical research ed) 1997;315(7109):629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy GH. Mendelian proportions in a mixed population. Science. 1908;28(706):49–50. doi: 10.1126/science.28.706.49. [DOI] [PubMed] [Google Scholar]
- 15.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. Journal of the National Cancer Institute. 2004;96(6):434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas DC, Clayton DG. Betting odds and genetic associations. Journal of the National Cancer Institute. 2004;96(6):421–3. doi: 10.1093/jnci/djh094. [DOI] [PubMed] [Google Scholar]
- 17.Kasperzyk JL, Chang ET, Birmann BM, Kraft P, Zheng T, Mueller NE. Nutrients and Genetic Variation Involved in One-Carbon Metabolism and Hodgkin Lymphoma Risk: A Population-based Case-Control Study. American Journal of Epidemiology. 2011 doi: 10.1093/aje/kwr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lourenco GJ, Neri IA, Sforni VC, Kameo R, Lorand-Metze I, Lima CS. Polymorphisms of glutathione S-transferase Mu 1, glutathione S-transferase theta 1 and glutathione S-transferase Pi 1 genes in Hodgkin's lymphoma susceptibility and progression. Leukemia & lymphoma. 2009;50(6):1005–9. doi: 10.1080/10428190902878455. [DOI] [PubMed] [Google Scholar]
- 19.El-Zein R, Monroy CM, Etzel CJ, Cortes AC, Xing Y, Collier AL, et al. Genetic polymorphisms in DNA repair genes as modulators of Hodgkin disease risk. Cancer. 2009;115(8):1651–9. doi: 10.1002/cncr.24205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monroy CM, Cortes AC, Lopez MS, D'Amelio AM, Jr, Etzel CJ, Younes A, et al. Hodgkin disease risk: role of genetic polymorphisms and gene-gene interactions in inflammation pathway genes. Molecular carcinogenesis. 2011;50(1):36–46. doi: 10.1002/mc.20688. [DOI] [PubMed] [Google Scholar]
- 21.Cozen W, Gill PS, Ingles SA, Masood R, Martinez-Maza O, Cockburn MG, et al. IL-6 levels and genotype are associated with risk of young adult Hodgkin lymphoma. Blood. 2004;103(8):3216–21. doi: 10.1182/blood-2003-08-2860. [DOI] [PubMed] [Google Scholar]
- 22.Chang ET, Birmann BM, Kasperzyk JL, Conti DV, Kraft P, Ambinder RF, et al. Polymorphic variation in NFKB1 and other aspirin-related genes and risk of Hodgkin lymphoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18(3):976–86. doi: 10.1158/1055-9965.EPI-08-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordano P, Lake A, Shield L, Taylor GM, Alexander FE, Taylor PR, et al. Effect of IL-6 promoter polymorphism on incidence and outcome in Hodgkin's lymphoma. Br J Haematol. 2005;128(4):493–5. doi: 10.1111/j.1365-2141.2004.05353.x. [DOI] [PubMed] [Google Scholar]
- 24.Yri OE, Ekstrom PO, Hilden V, Gaudernack G, Liestol K, Smeland EB, et al. Polymorphisms in genes encoding interleukin-10 and drug metabolizing enzymes GSTP1, GSTT1, GSTA1 and UGT1A1 influence risk and outcome in Hodgkin lymphoma. Leukemia & lymphoma. 2012;53(10):1934–44. doi: 10.3109/10428194.2012.682307. [DOI] [PubMed] [Google Scholar]
- 25.Munro LR, Johnston PW, Marshall NA, Canning SJ, Hewitt SG, Tveita K, et al. Polymorphisms in the interleukin-10 and interferon gamma genes in Hodgkin lymphoma. Leukemia & lymphoma. 2003;44(12):2083–8. doi: 10.1080/1042819031000119316. [DOI] [PubMed] [Google Scholar]
- 26.Hohaus S, Massini G, D'Alo F, Guidi F, Putzulu R, Scardocci A, et al. Association between glutathione S-transferase genotypes and Hodgkin's lymphoma risk and prognosis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9(9):3435–40. [PubMed] [Google Scholar]
- 27.Hohaus S, Di Ruscio A, Di Febo A, Massini G, D'Alo F, Guidi F, et al. Glutathione S-transferase P1 genotype and prognosis in Hodgkin's lymphoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(6):2175–9. doi: 10.1158/1078-0432.CCR-04-1250. [DOI] [PubMed] [Google Scholar]
- 28.Nieters A, Beckmann L, Deeg E, Becker N. Gene polymorphisms in Toll-like receptors, interleukin-10, and interleukin-10 receptor alpha and lymphoma risk. Genes and immunity. 2006;7(8):615–24. doi: 10.1038/sj.gene.6364337. [DOI] [PubMed] [Google Scholar]
- 29.Cunningham LM, Chapman C, Dunstan R, Bell MC, Joske DJ. Polymorphisms in the interleukin 10 gene promoter are associated with susceptibility to aggressive non-Hodgkin's lymphoma. Leukemia & lymphoma. 2003;44(2):251–5. doi: 10.1080/1042819021000035590. [DOI] [PubMed] [Google Scholar]
- 30.Sarmanova J, Benesova K, Gut I, Nedelcheva-Kristensen V, Tynkova L, Soucek P. Genetic polymorphisms of biotransformation enzymes in patients with Hodgkin's and non-Hodgkin's lymphomas. Human molecular genetics. 2001;10(12):1265–73. doi: 10.1093/hmg/10.12.1265. [DOI] [PubMed] [Google Scholar]
- 31.Ribrag V, Koscielny S, Casasnovas O, Cazeneuve C, Brice P, Morschhauser F, et al. Pharmacogenetic study in Hodgkin lymphomas reveals the impact of UGT1A1 polymorphisms on patient prognosis. Blood. 2009;113(14):3307–13. doi: 10.1182/blood-2008-03-148874. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz-Cosano J, Conesa-Zamora P, Gonzalez-Conejero R, Perez-Ceballos E, Martinez-Frances A, Vicente V, et al. Role of GSTT1 and M1 null genotypes as risk factors for B-cell lymphoma: influence of geographical factors and occupational exposure. Molecular carcinogenesis. 2012;51(6):508–13. doi: 10.1002/mc.20814. [DOI] [PubMed] [Google Scholar]
- 33.Lemos MC, Cabrita FJ, Silva HA, Vivan M, Placido F, Regateiro FJ. Genetic polymorphism of CYP2D6, GSTM1 and NAT2 and susceptibility to haematological neoplasias. Carcinogenesis. 1999;20(7):1225–9. doi: 10.1093/carcin/20.7.1225. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz-Cosano J, Torres-Moreno D, Conesa-Zamora P. Influence of polymorphisms in ERCC5, XPA and MTR DNA repair and synthesis genes in B-cell lymphoma risk. A case-control study in Spanish population. Journal of BUON : official journal of the Balkan Union of Oncology. 2013;18(2):486–90. [PubMed] [Google Scholar]
- 35.Timuragaoglu A, Dizlek S, Uysalgil N, Tosun O, Yamac K. Methylenetetrahydrofolate reductase C677T polymorphism in adult patients with lymphoproliferative disorders and its effect on chemotherapy. Annals of hematology. 2006;85(12):863–8. doi: 10.1007/s00277-006-0175-4. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez Ordonez AJ, Fernandez Carreira JM, Fernandez Alvarez CR, Martin L, Sanchez Garcia J, Medina Rodriguez JM, et al. Normal frequencies of the C677T genotypes on the methylenetetrahydrofolate reductase (MTHFR) gene among lymphoproliferative disorders but not in multiple myeloma. Leukemia & lymphoma. 2000;39(5–6):607–12. doi: 10.3109/10428190009113391. [DOI] [PubMed] [Google Scholar]
- 37.Deligezer U, Akisik EE, Yaman F, Erten N, Dalay N. MTHFR C677 T gene polymorphism in lymphoproliferative diseases. Journal of clinical laboratory analysis. 2006;20(2):37–41. doi: 10.1002/jcla.20103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoeft B, Becker N, Deeg E, Beckmann L, Nieters A. Joint effect between regular use of non-steroidal anti-inflammatory drugs, variants in inflammatory genes and risk of lymphoma. Cancer causes & control : CCC. 2008;19(2):163–73. doi: 10.1007/s10552-007-9082-9. [DOI] [PubMed] [Google Scholar]
- 39.Torres-Espindola LM, Velazquez-Cruz R, Falfan-Valencia R, Chavez-Pacheco JL, Salcedo-Vargas M, de Jesus Nambo-Lucio M, et al. Genetic polymorphism of tumor necrosis factor promoter region and susceptibility to develop Hodgkin lymphoma in a Mexican population. Leukemia & lymphoma. 2014;55(6):1295–9. doi: 10.3109/10428194.2013.842982. [DOI] [PubMed] [Google Scholar]
- 40.Ghesquières H, Dogan A, Link BK, Maurer MJ, Cunningham JM, Novak AJ, et al. FCGR2A and FCGR3A polymorphisms in classical Hodgkin lymphoma by Epstein–Barr virus status. Leukemia & lymphoma. 2013;54(11):2571–3. doi: 10.3109/10428194.2013.796048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monroy CM, Cortes AC, Lopez M, Rourke E, Etzel CJ, Younes A, et al. Hodgkin lymphoma risk: role of genetic polymorphisms and gene-gene interactions in DNA repair pathways. Molecular carcinogenesis. 2011;50(11):825–34. doi: 10.1002/mc.20747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Havranek O, Spacek M, Hubacek P, Mocikova H, Benesova K, Soucek P, et al. No association between the TP53 codon 72 polymorphism and risk or prognosis of Hodgkin and non-Hodgkin lymphoma. Leukemia research. 2011;35(8):1117–9. doi: 10.1016/j.leukres.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Broderick P, Cunningham D, Vijayakrishnan J, Cooke R, Ashworth A, Swerdlow A, et al. IRF4 polymorphism rs872071 and risk of Hodgkin lymphoma. British Journal of Haematology. 2010;148(3):413–5. doi: 10.1111/j.1365-2141.2009.07946.x. [DOI] [PubMed] [Google Scholar]
- 44.de Oliveira KB, Oda JM, Voltarelli JC, Nasser TF, Ono MA, Fujita TC, et al. CXCL12 rs1801157 polymorphism in patients with breast cancer, Hodgkin's lymphoma, and non-Hodgkin's lymphoma. Journal of clinical laboratory analysis. 2009;23(6):387–93. doi: 10.1002/jcla.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostrovsky O, Korostishevsky M, Levite I, Leiba M, Galski H, Vlodavsky I, et al. Association of heparanase gene (HPSE) single nucleotide polymorphisms with hematological malignancies. Leukemia. 2007;21(11):2296–303. doi: 10.1038/sj.leu.2404821. [DOI] [PubMed] [Google Scholar]
- 46.Wihlborg C, Sjoberg J, Intaglietta M, Axdorph U, Pisa EK, Pisa P. Tumour necrosis factor-alpha cytokine promoter gene polymorphism in Hodgkin's disease and chronic lymphocytic leukaemia. Br J Haematol. 1999;104(2):346–9. doi: 10.1046/j.1365-2141.1999.01176.x. [DOI] [PubMed] [Google Scholar]
- 47.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urayama KY, Jarrett RF, Hjalgrim H, Diepstra A, Kamatani Y, Chabrier A, et al. Genome-wide association study of classical Hodgkin lymphoma and Epstein-Barr virus status-defined subgroups. Journal of the National Cancer Institute. 2012;104(3):240–53. doi: 10.1093/jnci/djr516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cozen W, Timofeeva MN, Li D, Diepstra A, Hazelett D, Delahaye-Sourdeix M, et al. A meta-analysis of Hodgkin lymphoma reveals 19p13.3 TCF3 as a novel susceptibility locus. Nature communications. 2014;5 doi: 10.1038/ncomms4856. 3856. [DOI] [PMC free article] [PubMed] [Google Scholar]