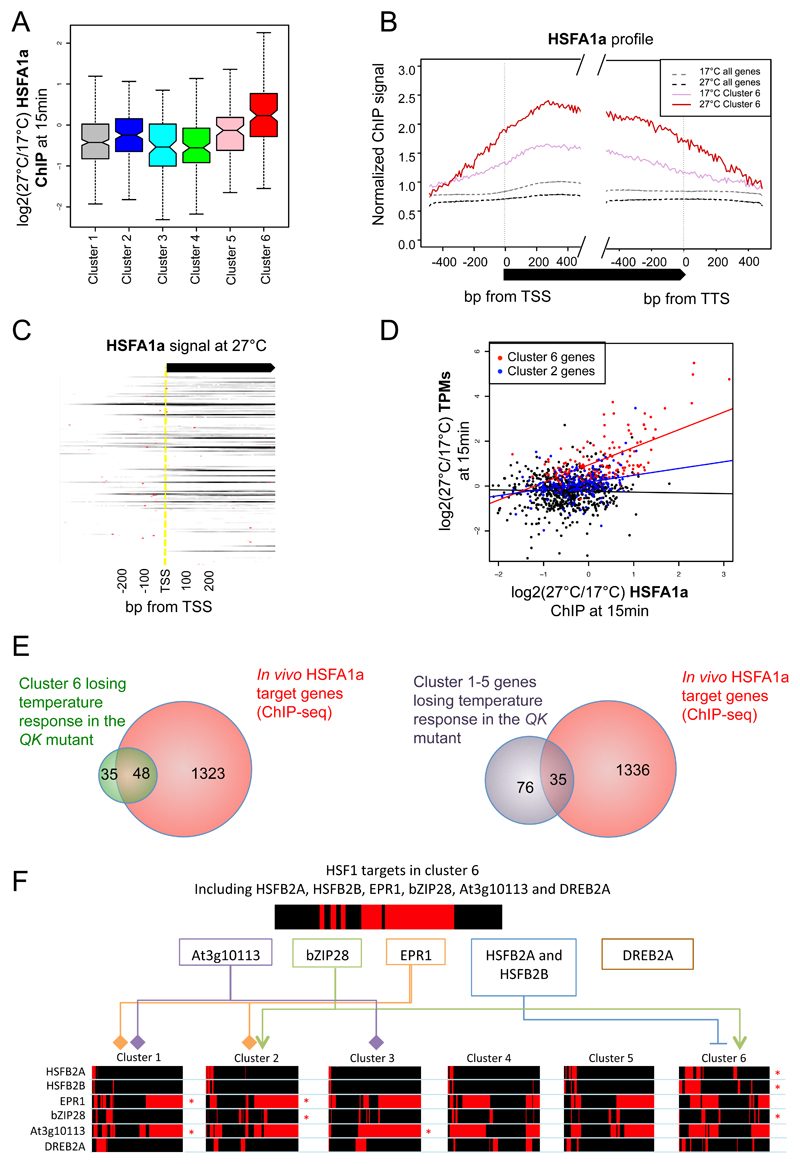
Figure 2. HSFA1a transcription factor binding is increasing at 27°C at the cluster 6 genes.
A Boxplot of changes for HSFA1a signal in the gene body between 17°C and 27°C after 15 min for genes in each temperature-responsive cluster. HSFA1a ChIP signal increases after temperature shift to 27°C specifically for cluster 6 genes (Wilcoxon test P-value < 2.2e-16 when comparing cluster 6 genes to genes in clusters 1-5). Non-overlapping notches indicate significant differences between populations’ medians.
B Genome-wide average HSFA1a binding profiles of HSFA1a show a higher HSFA1a occupancy at genes in cluster 6 (solid, pink) as compared to at 17°C (dotted, grey). The HSFA1a occupancy markedly increases after 15 min shift to 27°C (solid, red), as compared to 17°C (solid, pink),
C Gene-by-gene HSFA1a ChIP signal of cluster 6 genes at 27°C. Red dots indicate predicted HSEs from a known consensus motif.
D Correlation between changes in transcripts level (TPMs) and HSFA1a signal after 15min of shift from 17°C to 27°C. A strong positive correlation is observed between increase in transcripts level and HSFA1a signal for cluster 6 genes (red), as compared to an equal number of random genes from other temperature-responsive gene clusters (black) and cluster 2 genes (blue).
E Comparison of in vivo HSFA1a targets genes with genes losing temperature response in the QK hsfa1abde mutant that are either in the cluster 6 (left) or in clusters 1-5 (right). Of all the 147 temperature-rapid-response genes (cluster 6), 83 (~56%) become temperature-irresponsive in the QK mutant as compared to the TK wild-type. Temperature responsiveness is defined by log2(TPM 27°C 15 min/TPM 17°C 15 mins) >= 0.5, or the TPM 27°C at 15 mins is less than 2 (undetectable), while TPM 17°C at 15 mins is not. Of these 83 genes, 48 (~58%) are shown to be direct targets of HSFA1a (Fisher’s exact test P-value < 2.2e-16), signifying the crucial role of HSFA1a and other TFs in the HSFA families in transcriptional regulation temperature rapid responsive genes. For other temperature responsive genes (cluster 1-5), 35 out of 111 (~32%) of genes losing temperature-responsiveness are direct targets of HSFA1a. Note that there are 1,371 genes (~5%) predicted as targets of HSFA1a in the Arabidopsis genome.
F Representation of the transcriptional cascade in the clusters 1-6 genes (bottom) by the TFs that are in the cluster 6 and are HSFA1a targets (top). The direct targets of these TFs were determined based on the available DAP-seq dataset (O’Malley et al., 2016) and are represented in red stripes for each cluster (bottom). The linkers and asterisks indicates the temperature-responsive clusters whose members are significantly enriched in target genes of these temperature-responsive TFs. Known positive and negative regulatory relationships are indicated by arrows and blunt arrows, respectively. HSFB2A/B, EPR1, bZIP28, DREB2A and At3g10113 TFs in the cluster 6 are transcriptionally activated by HSFA1a, and they themselves regulate downstream temperature responsive targets.