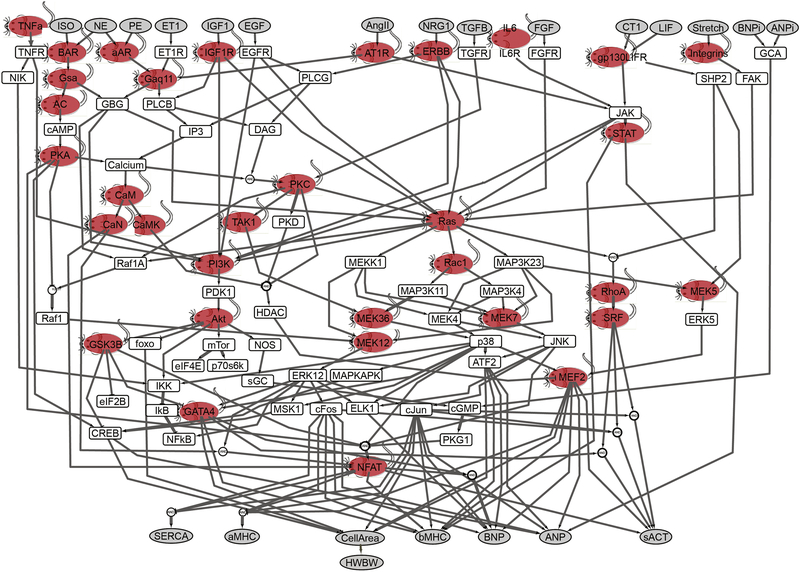
Figure 1. Transgenic mouse models developed to overexpress many of the species in the cardiac hypertrophy signaling network model.
The model consists of 106 species and 192 reactions. Model inputs and outputs are shown in gray. This model focused on neonatal rat ventricular myocytes for initial implementation using a normalized-Hill differential equation approach [2]. Fifty-two transgenic mouse models representing 34 of the species were identified in the literature for this analysis (red mice, see supplemental Table S5). ISO, isoproterenol; NE, norepinephrine; ET1, endothelin-1; IGF1, insulin-like growth factor 1; AngII, angiotensin II; NGR1, neuregulin 1; IL6, interleukin 6; FGF, fibroblast growth factor; CT1, cardiotropin 1; BNP, brain natriuretic peptide; ANP, atrial natriuretic peptide; SERCA, sarcoplasmic reticulum ATPase; α-MHC, α- myosin heavy chain; sACT, skeletal α-actin; CREB, cAMP-responsive element-binding protein; EGFR, EGF receptor; FGFR, FGF receptor; IP3, inositol 1,4,5-trisphosphate; β-AR, β-adrenergic receptor; α-AR, α-adrenergic receptor; NOS, nitric oxide synthase; NIK, NFκB-inducing kinase; AC, adenylyl cyclase; DAG, diacylglycerol; PLC, phospholipase C; FAK, focal adhesion kinase; SRF, serum response factor; GCA, guanylyl cyclase subtype A.