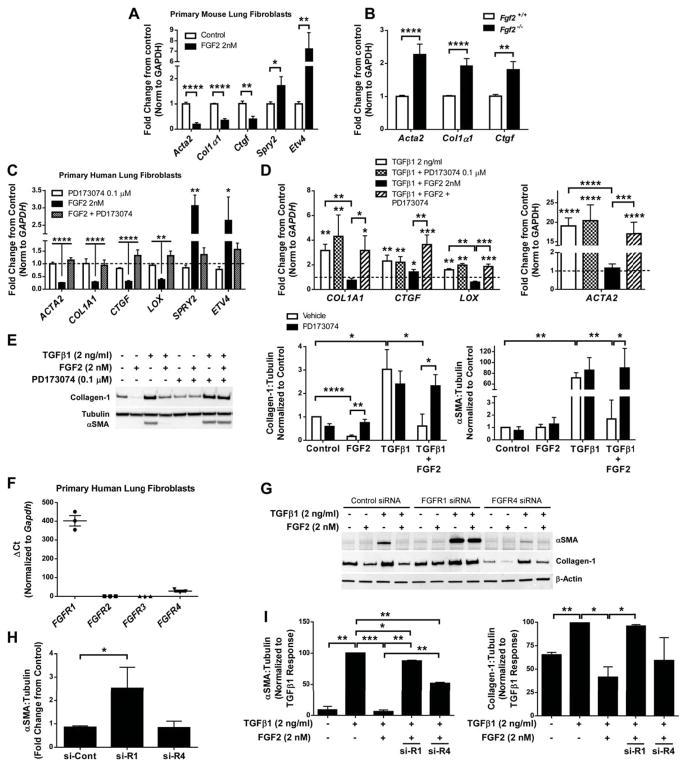
Figure 4.
Recombinant FGF2 inhibits expression of pro-fibrotic genes in primary cultured lung fibroblasts in an FGFR1-dependent manner. (A) Wild-type mouse lung fibroblasts were serum-starved overnight and treated with recombinant FGF2 (2 nM). 48 hours after treatment total RNA was collected and analyzed via qRT-PCR for expression of Acta2, Col1α1, Ctgf, Spry2, and Etv4. (B) Total RNA was collected from primary lung fibrobasts isolated from Fgf2 +/+ and Fgf2−/− mice, and qRT-PCR was performed for Acta2, Col1α1, and Ctgf. ΔCt values were normalized to Gapdh and expressed as fold change from untreated controls. (C) Primary HLFs were serum-starved overnight and treated with FGF2 (2 nM) and/or PD173074 (0.1 μM) for 48 hours prior to total RNA collection. qRT-PCR was performed for ACTA2, COL1A1, CTGF, LOX, SPRY2, and ETV4. (D) Serum-starved primary HLFs were treated with TGFβ1 (2 ng/ml) or FGF2+TGFβ1 +/− PD173074 (0.1 μM) for 48 hours. qRT-PCR was performed for COL1A1, LOX, and ACTA2. ΔCt values for C and D were normalized to GAPDH and expressed as fold change from untreated controls (represented as dashed line). (E) Total protein was collected from primary HLFs treated with FGF2, TGFβ1, or FGF2+TGFβ1 +/− PD173074 for 48 hours. Collagen-1 and αSMA were measured via western blot, normalized to tubulin, and expressed as fold change from untreated controls. (F) RNA from untreated early passage HLFs was analyzed via qRT-PCR for FGFR1, FGFR2, FGFR3, and FGFR4. ΔCt values were normalized to GAPDH. (G) HLFs were transfected with control siRNA, FGFR1 siRNA, or FGFR4 siRNA, and treated with FGF2, TGFβ1, or TGFβ1+FGF2. Total protein was analyzed via western blot for αSMA, collagen-1, and beta-actin. Densitometric data was quantified and is expressed as fold change from control siRNA (H) and % of TGFβ1 response (I). “ns” = not significant. * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.005, **** indicates p < 0.001 using an unpaired 2-way t-test.