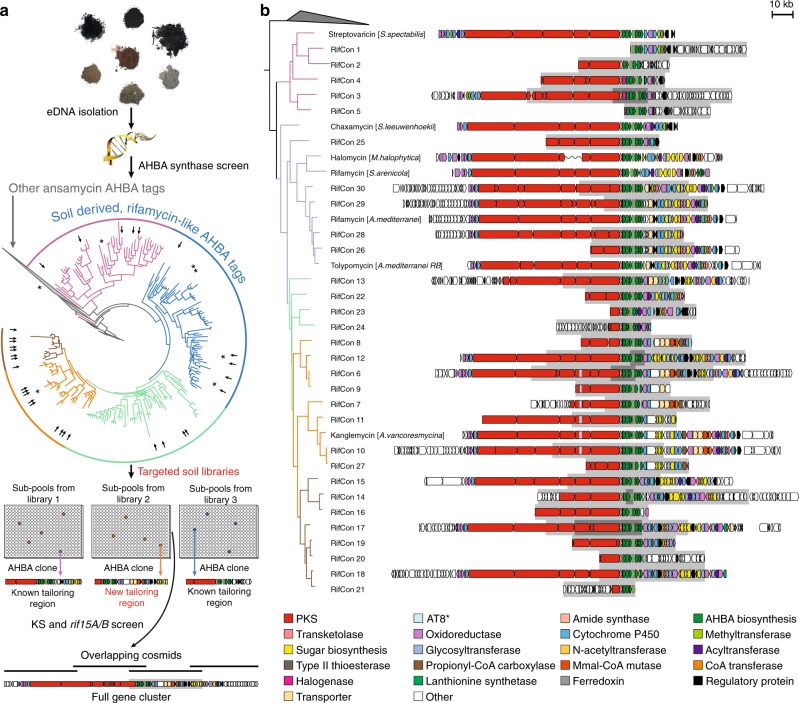
Fig. 2.
Sequence-based screen for rifamycin congener gene clusters. a Screening overview. DNA isolated from ~1500 soils was screened for the presence of AHBA synthase genes by PCR using degenerate primers. Sequence tags generated in this screen were used to construct a phylogenetic tree, onto which AHBA synthase reference sequences from known rifamycin congener gene clusters were mapped (marked with asterisks). Large, distinct clades in the phylogenetic tree are shown in different colors. Metagenomic DNA cosmid libraries were generated from soils that contained AHBA sequence tags that spanned all AHBA clades predicted to be associated with rifamycin congener gene clusters. To facilitate the recovery of individual clones containing gene clusters of interest, each metagenomic library was expanded to contain >20,000,000 unique eDNA cosmids and formatted as smaller subpools of between 20,000 and 60,000 unique cosmid clones per sub-pool. Primary clones (those containing an AHBA synthase gene) were recovered from AHBA positive subpools using a PCR dilution method and degenerate AHBA synthase primers. The same approach, but with degenerate primers targeting PKS ketosynthase (KS) domains and the rif15A/15B tailoring genes, was used to recover regions of the pathways that flank those found on the primary clone. AHBA sequence tags corresponding with primary clones that were targeted for recovery are indicated with arrows on the phylogenetic tree. b Summary of rifamycin congener gene clusters recovered from the soil metagenomes. Portions of the gene clusters found on primary clones are shown on a gray background