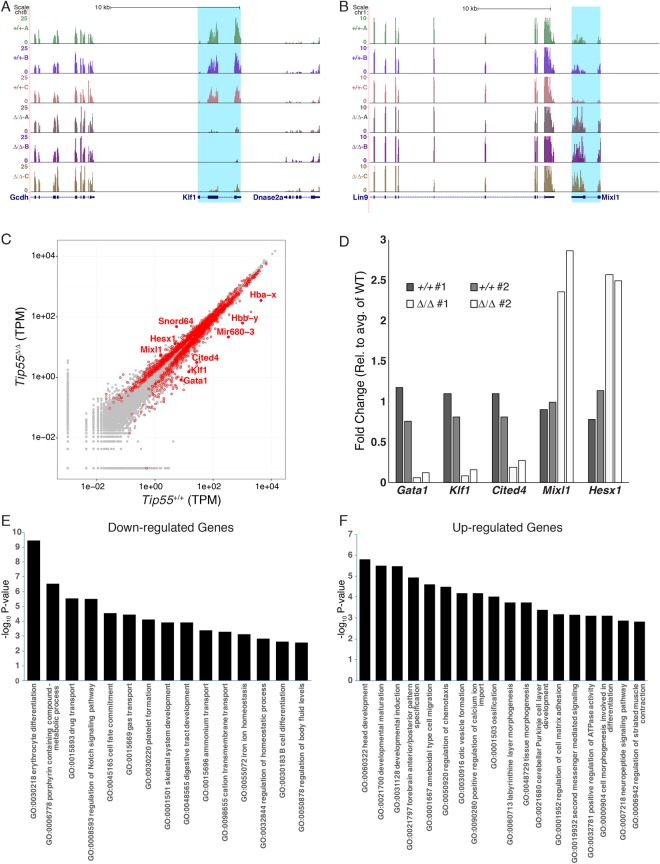
Figure 5.
Genes necessary for organogenesis are misregulated in Tip55 mutant embryos. (A,B) Browser tracks of developmental regulators (highlighted in blue) downregulated (A) or upregulated (B) in Tip55Δ/Δ embryos (Δ/Δ) relative to Tip55+/+ embryos (+/+). Three biological replicate RNA-seq datasets (normalized for read number) for each geneotype were performed. (C) Average transcripts per million (TPM) for each genotype are shown on a log-log scale with genes significantly differentially expressed in Tip55Δ/Δ embryos (posterior probability of differential expression; PPDE > 0.95) highlighted with red circles. Several genes of interest labeled with solid red dots with gene names shown. (D) RT-qPCR validation of indicated genes from Tip55+/+ (+/+) or Tip55Δ/Δ (Δ/Δ) E8.5 embryos. Genes were selected based on differential expression (up or downregulation) in RNA-seq experiments. Expression levels in biological duplicate Tip55+/+ or Tip55Δ/Δ embryos are plotted individually, relative to the average of the Tip55+/+ (which is set to 1). (E,F) Significantly enriched gene ontology (GO) categories for genes down-regulated or up-regulated significantly (PPDE > 0.95) and |log2 (Fold Change)| >0.6 are depicted in (A) and (B), respectively.